Diversity and evolution of the emerging Pandoraviridae family
- PMID: 29891839
- PMCID: PMC5995976
- DOI: 10.1038/s41467-018-04698-4
Diversity and evolution of the emerging Pandoraviridae family
Abstract
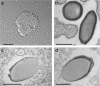
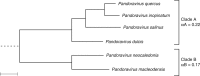
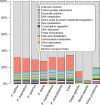
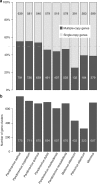
With DNA genomes reaching 2.5 Mb packed in particles of bacterium-like shape and dimension, the first two Acanthamoeba-infecting pandoraviruses remained up to now the most complex viruses since their discovery in 2013. Our isolation of three new strains from distant locations and environments is now used to perform the first comparative genomics analysis of the emerging worldwide-distributed Pandoraviridae family. Thorough annotation of the genomes combining transcriptomic, proteomic, and bioinformatic analyses reveals many non-coding transcripts and significantly reduces the former set of predicted protein-coding genes. Here we show that the pandoraviruses exhibit an open pan-genome, the enormous size of which is not adequately explained by gene duplications or horizontal transfers. As most of the strain-specific genes have no extant homolog and exhibit statistical features comparable to intergenic regions, we suggest that de novo gene creation could contribute to the evolution of the giant pandoravirus genomes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

