Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants
- PMID: 29892050
- PMCID: PMC5995836
- DOI: 10.1038/s41467-018-04109-8
Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants
Abstract
Prostate cancer is a polygenic disease with a large heritable component. A number of common, low-penetrance prostate cancer risk loci have been identified through GWAS. Here we apply the Bayesian multivariate variable selection algorithm JAM to fine-map 84 prostate cancer susceptibility loci, using summary data from a large European ancestry meta-analysis. We observe evidence for multiple independent signals at 12 regions and 99 risk signals overall. Only 15 original GWAS tag SNPs remain among the catalogue of candidate variants identified; the remainder are replaced by more likely candidates. Biological annotation of our credible set of variants indicates significant enrichment within promoter and enhancer elements, and transcription factor-binding sites, including AR, ERG and FOXA1. In 40 regions at least one variant is colocalised with an eQTL in prostate cancer tissue. The refined set of candidate variants substantially increase the proportion of familial relative risk explained by these known susceptibility regions, which highlights the importance of fine-mapping studies and has implications for clinical risk profiling.
Conflict of interest statement
The authors declare no competing interests.
Figures
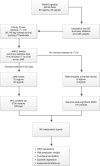
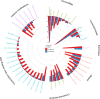

References
Publication types
MeSH terms
Grants and funding
- 18504/CRUK_/Cancer Research UK/United Kingdom
- 17528/CRUK_/Cancer Research UK/United Kingdom
- 21332/CRUK_/Cancer Research UK/United Kingdom
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- G1000143/MRC_/Medical Research Council/United Kingdom
- 14136/CRUK_/Cancer Research UK/United Kingdom
- G0401527/MRC_/Medical Research Council/United Kingdom
- 15007/CRUK_/Cancer Research UK/United Kingdom
- 19170/CRUK_/Cancer Research UK/United Kingdom
- K07 CA187546/CA/NCI NIH HHS/United States
- 16491/CRUK_/Cancer Research UK/United Kingdom
- U10 CA037429/CA/NCI NIH HHS/United States
- 13232/CRUK_/Cancer Research UK/United Kingdom
- 16565/CRUK_/Cancer Research UK/United Kingdom
- UG1 CA189974/CA/NCI NIH HHS/United States
- UM1 CA182883/CA/NCI NIH HHS/United States
- MC_UU_00002/9/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

