The human heart contains distinct macrophage subsets with divergent origins and functions
- PMID: 29892064
- PMCID: PMC6082687
- DOI: 10.1038/s41591-018-0059-x
The human heart contains distinct macrophage subsets with divergent origins and functions
Abstract
Paradigm-shifting studies in the mouse have identified tissue macrophage heterogeneity as a critical determinant of immune responses. In contrast, surprisingly little is known regarding macrophage heterogeneity in humans. Macrophages within the mouse heart are partitioned into CCR2- and CCR2+ subsets with divergent origins, repopulation mechanisms, and functions. Here, we demonstrate that the human myocardium also contains distinct subsets of CCR2- and CCR2+ macrophages. Analysis of sex-mismatched heart transplant recipients revealed that CCR2- macrophages are a tissue-resident population exclusively replenished through local proliferation, whereas CCR2+ macrophages are maintained through monocyte recruitment and proliferation. Moreover, CCR2- and CCR2+ macrophages have distinct functional properties, analogous to reparative CCR2- and inflammatory CCR2+ macrophages in the mouse heart. Clinically, CCR2+ macrophage abundance is associated with left ventricular remodeling and systolic function in heart failure patients. Collectively, these observations provide initial evidence for the functional importance of macrophage heterogeneity in the human heart.
Conflict of interest statement
The authors have no competing financial interests to disclose.
Figures

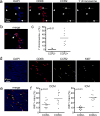
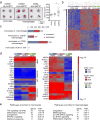
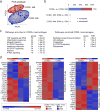

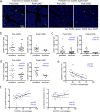
Comment in
-
Macrophages that fix or break the heart.Sci Immunol. 2018 Jul 6;3(25):eaau2828. doi: 10.1126/sciimmunol.aau2828. Sci Immunol. 2018. PMID: 29980622
-
Cellular origin of human cardiac macrophage populations.Nat Med. 2018 Aug;24(8):1091-1092. doi: 10.1038/s41591-018-0143-2. Nat Med. 2018. PMID: 30082862 No abstract available.
References
-
- Volkman A, Chang NC, Strausbauch PH, Morahan PS. Differential effects of chronic monocyte depletion on macrophage populations. Lab Invest. 1983;49:291–298. - PubMed
-
- Sawyer RT, Strausbauch PH, Volkman A. Resident macrophage proliferation in mice depleted of blood monocytes by strontium-89. Lab Invest. 1982;46:165–170. - PubMed
-
- Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL135121/HL/NHLBI NIH HHS/United States
- R01 HL131908/HL/NHLBI NIH HHS/United States
- K08 HL123519/HL/NHLBI NIH HHS/United States
- R01 HL138466/HL/NHLBI NIH HHS/United States
- R35 HL139598/HL/NHLBI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- I01 BX002730/BX/BLRD VA/United States
- R01 HL139714/HL/NHLBI NIH HHS/United States
- P01 AI116501/AI/NIAID NIH HHS/United States
- R01 HL125655/HL/NHLBI NIH HHS/United States
- R01 HL094601/HL/NHLBI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

