Plasticity in binding confers selectivity in ligand-induced protein degradation
- PMID: 29892083
- PMCID: PMC6202246
- DOI: 10.1038/s41589-018-0055-y
Plasticity in binding confers selectivity in ligand-induced protein degradation
Abstract
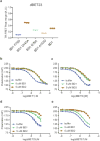
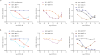
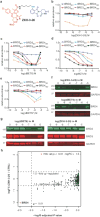
Heterobifunctional small-molecule degraders that induce protein degradation through ligase-mediated ubiquitination have shown considerable promise as a new pharmacological modality. However, we currently lack a detailed understanding of the molecular basis for target recruitment and selectivity, which is critically required to enable rational design of degraders. Here we utilize a comprehensive characterization of the ligand-dependent CRBN-BRD4 interaction to demonstrate that binding between proteins that have not evolved to interact is plastic. Multiple X-ray crystal structures show that plasticity results in several distinct low-energy binding conformations that are selectively bound by ligands. We demonstrate that computational protein-protein docking can reveal the underlying interprotein contacts and inform the design of a BRD4 selective degrader that can discriminate between highly homologous BET bromodomains. Our findings that plastic interprotein contacts confer selectivity for ligand-induced protein dimerization provide a conceptual framework for the development of heterobifunctional ligands.
Conflict of interest statement
E.S.F. is a member of the scientific advisory board of C4 Therapeutics and a consultant to Novartis Pharmaceuticals. N.S.G. is a founder and scientific advisory board member of C4 Therapeutics. J.E.B. is an executive and shareholder of Novartis Pharmaceuticals.
Figures
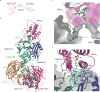


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

