Endophilin A1 Promotes Actin Polymerization in Dendritic Spines Required for Synaptic Potentiation
- PMID: 29892212
- PMCID: PMC5985315
- DOI: 10.3389/fnmol.2018.00177
Endophilin A1 Promotes Actin Polymerization in Dendritic Spines Required for Synaptic Potentiation
Abstract
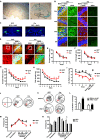
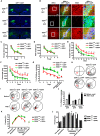
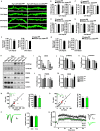
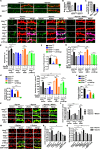
Endophilin A1 is a member of the N-BAR domain-containing endophilin A protein family that is involved in membrane dynamics and trafficking. At the presynaptic terminal, endophilin As participate in synaptic vesicle recycling and autophagosome formation. By gene knockout studies, here we report that postsynaptic endophilin A1 functions in synaptic plasticity. Ablation of endophilin A1 in the hippocampal CA1 region of mature mouse brain impairs long-term spatial and contextual fear memory. Its loss in CA1 neurons postsynaptic of the Schaffer collateral pathway causes impairment in their AMPA-type glutamate receptor-mediated synaptic transmission and long-term potentiation. In KO neurons, defects in the structural and functional plasticity of dendritic spines can be rescued by overexpression of endophilin A1 but not A2 or A3. Further, endophilin A1 promotes actin polymerization in dendritic spines during synaptic potentiation. These findings reveal a physiological role of endophilin A1 distinct from that of other endophilin As at the postsynaptic site.
Keywords: AMPAR; actin polymerization; dendritic spine; endophilin A1; learning and memory; structural plasticity; synaptic potentiation; synaptic transmission.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

