Biochemical Comparison of dsRNA Degrading Nucleases in Four Different Insects
- PMID: 29892232
- PMCID: PMC5985623
- DOI: 10.3389/fphys.2018.00624
Biochemical Comparison of dsRNA Degrading Nucleases in Four Different Insects
Abstract
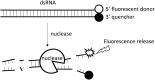
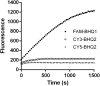
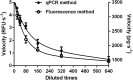
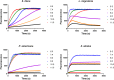
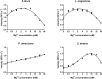
Double stranded RNAs (dsRNA) degrading nuclease is responsible for the rapid degradation of dsRNA molecules, and thus accounts for variations in RNA interference (RNAi) efficacy among insect species. Here, the biochemical properties and tissue-specific activities of dsRNA degrading nucleases in four insects (Spodoptera litura, Locusta migratoria, Periplaneta americana, and Zophobas atratus) from different orders were characterized using a modified assay method. The results revealed that all insect dsRNA degrading nucleases tested showed high activity in alkaline environments at optimal Mg2+ concentrations and elevated temperatures. We also found that enzymes from different insects varied in terms of their optimal reaction conditions and kinetic parameters. Whole body enzyme activity differed dramatically between insect species, although enzymes with higher substrate affinities (lower Km) were usually balanced by a smaller Vmax to maintain a proper level of degradative capacity. Furthermore, enzyme activities varied significantly between the four tested tissues (whole body, gut, hemolymph, and carcass) of the insect species. All the insects tested showed several hundred-fold higher dsRNA degrading activity in their gut than in other tissues. Reaction environment analysis demonstrated that physiological conditions in the prepared gut fluid and serum of different insects were not necessarily optimal for dsRNA degrading nuclease activity. Our data describe the biochemical characteristics and tissue distributions of dsRNA degrading activities in various insects, not only explaining why oral delivery of dsRNA often produces lower RNAi effects than injection of dsRNA, but also suggesting that dsRNA-degrading activities are regulated by physiological conditions. These results allow for a better understanding of the properties of dsRNA degrading nucleases, and will aid in the development of successful RNAi strategies in insects.
Keywords: RNA interference; dsRNA degrading enzyme; dsRNase; fluorescence; nuclease.
Figures



References
-
- Arimatsu Y., Furuno T., Sugimura Y., Togoh M., Ishihara R., Tokizane M., et al. (2007a). Purification and properties of double-stranded RNA-degrading nuclease, dsRNase, from the digestive juice of the silkworm, Bombyx mori. J. Insect Biotechnol. Sericol. 76 57–62.
LinkOut - more resources
Full Text Sources
Other Literature Sources

