A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses
- PMID: 29892288
- PMCID: PMC5985295
- DOI: 10.3389/fimmu.2018.01135
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses
Abstract
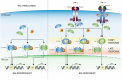
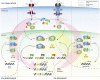
Interferon (IFN)-I and IFN-II both induce IFN-stimulated gene (ISG) expression through Janus kinase (JAK)-dependent phosphorylation of signal transducer and activator of transcription (STAT) 1 and STAT2. STAT1 homodimers, known as γ-activated factor (GAF), activate transcription in response to all types of IFNs by direct binding to IFN-II activation site (γ-activated sequence)-containing genes. Association of interferon regulatory factor (IRF) 9 with STAT1-STAT2 heterodimers [known as interferon-stimulated gene factor 3 (ISGF3)] or with STAT2 homodimers (STAT2/IRF9) in response to IFN-I, redirects these complexes to a distinct group of target genes harboring the interferon-stimulated response element (ISRE). Similarly, IRF1 regulates expression of ISGs in response to IFN-I and IFN-II by directly binding the ISRE or IRF-responsive element. In addition, evidence is accumulating for an IFN-independent and -dependent role of unphosphorylated STAT1 and STAT2, with or without IRF9, and IRF1 in basal as well as long-term ISG expression. This review provides insight into the existence of an intracellular amplifier circuit regulating ISG expression and controlling long-term cellular responsiveness to IFN-I and IFN-II. The exact timely steps that take place during IFN-activated feedback regulation and the control of ISG transcription and long-term cellular responsiveness to IFN-I and IFN-II is currently not clear. Based on existing literature and our novel data, we predict the existence of a multifaceted intracellular amplifier circuit that depends on unphosphorylated and phosphorylated ISGF3 and GAF complexes and IRF1. In a combinatorial and timely fashion, these complexes mediate prolonged ISG expression and control cellular responsiveness to IFN-I and IFN-II. This proposed intracellular amplifier circuit also provides a molecular explanation for the existing overlap between IFN-I and IFN-II activated ISG expression.
Keywords: JAK/signal transducer and activator of transcription signaling pathway; antiviral activity; interferon; interferon regulatory factor 1; interferon-stimulated gene factor 3; signal transducer and activator of transcriptions; transcriptional regulation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

