The polyphyly of Plasmodium: comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict
- PMID: 29892372
- PMCID: PMC5990803
- DOI: 10.1098/rsos.171780
The polyphyly of Plasmodium: comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict
Abstract
The evolutionary relationships among the apicomplexan blood pathogens known as the malaria parasites (order Haemosporida), some of which infect nearly 200 million humans each year, has remained a vexing phylogenetic problem due to limitations in taxon sampling, character sampling and the extreme nucleotide base composition biases that are characteristic of this clade. Previous phylogenetic work on the malaria parasites has often lacked sufficient representation of the broad taxonomic diversity within the Haemosporida or the multi-locus sequence data needed to resolve deep evolutionary relationships, rendering our understanding of haemosporidian life-history evolution and the origin of the human malaria parasites incomplete. Here we present the most comprehensive phylogenetic analysis of the malaria parasites conducted to date, using samples from a broad diversity of vertebrate hosts that includes numerous enigmatic and poorly known haemosporidian lineages in addition to genome-wide multi-locus sequence data. We find that if base composition differences were corrected for during phylogenetic analysis, we recovered a well-supported topology indicating that the evolutionary history of the malaria parasites was characterized by a complex series of transitions in life-history strategies and host usage. Notably we find that Plasmodium, the malaria parasite genus that includes the species of human medical concern, is polyphyletic with the life-history traits characteristic of this genus having evolved in a dynamic manner across the phylogeny. We find support for multiple instances of gain and loss of asexual proliferation in host blood cells and production of haemozoin pigment, two traits that have been used for taxonomic classification as well as considered to be important factors for parasite virulence and used as drug targets. Lastly, our analysis illustrates the need for a widespread reassessment of malaria parasite taxonomy.
Keywords: Plasmodium; base composition bias; malaria; phylogeny; polyphyly.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
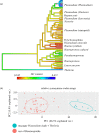

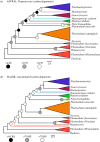

References
-
- Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. 1996. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 83, 141–152. (doi:10.1016/S0166-6851(96)02755-7) - DOI - PubMed
-
- Suzuki Y, Gojobori T. 1997. The origin and evolution of Ebola and Marburg viruses. Mol. Biol. Evol. 14, 800–806. (doi:10.1093/oxfordjournals.molbev.a025820) - DOI - PubMed
-
- Gao F, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441. (doi:10.1038/17130) - DOI - PubMed
-
- Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. 2001. Human immunodeficiency virus: phylogeny and the origin of HIV-1. Nature 410, 1047–1048. (doi:10.1038/35074179) - DOI - PubMed
-
- Zwickl DJ, Hillis DM. 2002. Increased taxon sampling greatly reduces phylogenetic error. Sys. Biol. 51, 588–598. (doi:10.1080/10635150290102339) - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
