Toll-Like Receptor Evolution in Birds: Gene Duplication, Pseudogenization, and Diversifying Selection
- PMID: 29893911
- PMCID: PMC6107061
- DOI: 10.1093/molbev/msy119
Toll-Like Receptor Evolution in Birds: Gene Duplication, Pseudogenization, and Diversifying Selection
Abstract
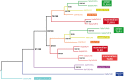
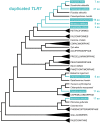
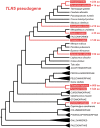
Toll-like receptors (TLRs) are key sensor molecules in vertebrates triggering initial phases of immune responses to pathogens. The avian TLR family typically consists of ten receptors, each adapted to distinct ligands. To understand the complex evolutionary history of each avian TLR, we analyzed all members of the TLR family in the whole genome assemblies and target sequence data of 63 bird species covering all major avian clades. Our results indicate that gene duplication events most probably occurred in TLR1 before synapsids diversified from sauropsids. Unlike mammals, ssRNA-recognizing TLR7 has duplicated independently in several avian taxa, while flagellin-sensing TLR5 has pseudogenized multiple times in bird phylogeny. Our analysis revealed stronger positive, diversifying selection acting in TLR5 and the three-domain TLRs (TLR10 [TLR1A], TLR1 [TLR1B], TLR2A, TLR2B, TLR4) that face the extracellular space and bind complex ligands than in single-domain TLR15 and endosomal TLRs (TLR3, TLR7, TLR21). In total, 84 out of 306 positively selected sites were predicted to harbor substitutions dramatically changing the amino acid physicochemical properties. Furthermore, 105 positively selected sites were located in the known functionally relevant TLR regions. We found evidence for convergent evolution acting between birds and mammals at 54 of these sites. Our comparative study provides a comprehensive insight into the evolution of avian TLR genetic variability. Besides describing the history of avian TLR gene gain and gene loss, we also identified candidate positions in the receptors that have been likely shaped by direct molecular host-pathogen coevolutionary interactions and most probably play key functional roles in birds.
Keywords: adaptive evolution; amino acid physicochemical properties; convergence; pattern recognition receptors; positive selection; pseudogene.
© The Author(s) 2018. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Gene duplication and adaptive evolution of Toll-like receptor genes in birds.Dev Comp Immunol. 2021 Jun;119:103990. doi: 10.1016/j.dci.2020.103990. Epub 2021 Jan 8. Dev Comp Immunol. 2021. PMID: 33422554
-
Molecular evolution of the vertebrate TLR1 gene family--a complex history of gene duplication, gene conversion, positive selection and co-evolution.BMC Evol Biol. 2011 May 28;11:149. doi: 10.1186/1471-2148-11-149. BMC Evol Biol. 2011. PMID: 21619680 Free PMC article.
-
Molecular evolution of the toll-like receptor multigene family in birds.Mol Biol Evol. 2011 May;28(5):1703-15. doi: 10.1093/molbev/msq351. Epub 2011 Jan 14. Mol Biol Evol. 2011. PMID: 21239391
-
Avian toll-like receptors.Cell Tissue Res. 2011 Jan;343(1):121-30. doi: 10.1007/s00441-010-1026-0. Epub 2010 Sep 1. Cell Tissue Res. 2011. PMID: 20809414 Review.
-
Toll-like receptors in bony fish: from genomics to function.Dev Comp Immunol. 2011 Dec;35(12):1263-72. doi: 10.1016/j.dci.2011.03.006. Epub 2011 Mar 15. Dev Comp Immunol. 2011. PMID: 21414346 Review.
Cited by
-
A new method for ecologists to estimate heterozygote excess and deficit for multi-locus gene families.Ecol Evol. 2024 Jul 23;14(7):e11561. doi: 10.1002/ece3.11561. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39045501 Free PMC article.
-
Cannabinoid receptor 2 evolutionary gene loss makes parrots more susceptible to neuroinflammation.Proc Biol Sci. 2022 Dec 14;289(1988):20221941. doi: 10.1098/rspb.2022.1941. Epub 2022 Dec 7. Proc Biol Sci. 2022. PMID: 36475439 Free PMC article.
-
Balancing Selection Drives the Maintenance of Genetic Variation in Drosophila Antimicrobial Peptides.Genome Biol Evol. 2019 Sep 1;11(9):2691-2701. doi: 10.1093/gbe/evz191. Genome Biol Evol. 2019. PMID: 31504505 Free PMC article.
-
Exploring the Papillomaviral Proteome to Identify Potential Candidates for a Chimeric Vaccine against Cervix Papilloma Using Immunomics and Computational Structural Vaccinology.Viruses. 2019 Jan 15;11(1):63. doi: 10.3390/v11010063. Viruses. 2019. PMID: 30650527 Free PMC article.
-
Comparative Genomics of the Waterfowl Innate Immune System.Mol Biol Evol. 2022 Aug 3;39(8):msac160. doi: 10.1093/molbev/msac160. Mol Biol Evol. 2022. PMID: 35880574 Free PMC article.
References
-
- Alcaide M, Edwards SV.. 2011. Molecular evolution of the Toll-like receptor multigene family in birds. Mol Biol Evol. 285:1703–1715. - PubMed
-
- Arbour N, Lorenz E, Schutte B, Zabner J, Kline J, Jones M, Frees K, Watt J, Schwartz D.. 2000. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 252:187–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

