Microfluidic chambers using fluid walls for cell biology
- PMID: 29895687
- PMCID: PMC6042120
- DOI: 10.1073/pnas.1805449115
Microfluidic chambers using fluid walls for cell biology
Abstract
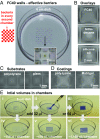
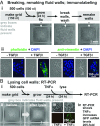
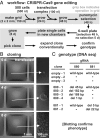
Many proofs of concept have demonstrated the potential of microfluidics in cell biology. However, the technology remains inaccessible to many biologists, as it often requires complex manufacturing facilities (such as soft lithography) and uses materials foreign to cell biology (such as polydimethylsiloxane). Here, we present a method for creating microfluidic environments by simply reshaping fluids on a substrate. For applications in cell biology, we use cell media on a virgin Petri dish overlaid with an immiscible fluorocarbon. A hydrophobic/fluorophilic stylus then reshapes the media into any pattern by creating liquid walls of fluorocarbon. Microfluidic arrangements suitable for cell culture are made in minutes using materials familiar to biologists. The versatility of the method is demonstrated by creating analogs of a common platform in cell biology, the microtiter plate. Using this vehicle, we demonstrate many manipulations required for cell culture and downstream analysis, including feeding, replating, cloning, cryopreservation, lysis plus RT-PCR, transfection plus genome editing, and fixation plus immunolabeling (when fluid walls are reconfigured during use). We also show that mammalian cells grow and respond to stimuli normally, and worm eggs develop into adults. This simple approach provides biologists with an entrée into microfluidics.
Keywords: fluid walls; interfacial tension; microfluidics; sessile drops; tissue culture.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: Oxford University Innovation—the technology transfer company of the University of Oxford—has filed provisional patent applications on behalf of C.S., A.F., P.R.C., and E.J.W. partly based on this study. A.F., H.W, P.R.C., and E.J.W. each hold equity, or rights to equity, in Iota Sciences Ltd., a company that is exploiting this technology. Iota Sciences Ltd. provides a scholarship for C.S. and partially funds salaries and research of A.F., A.N.T., H.W., and P.A.W.
Figures



References
-
- Ng K, et al. Paper-based cell culture platform and its emerging biomedical applications. Mater Today. 2017;20:32–44.
-
- Kim S-H, Lee GH, Park JW. Microwell fabrication methods and applications for cellular studies. Biomed Eng Lett. 2013;3:131–137.
-
- Liberski AR, Delaney JT, Jr, Schubert US. “One cell-one well”: A new approach to inkjet printing single cell microarrays. ACS Comb Sci. 2011;13:190–195. - PubMed
-
- Hoffmann J, Trotter M, von Stetten F, Zengerle R, Roth G. Solid-phase PCR in a picowell array for immobilizing and arraying 100,000 PCR products to a microscope slide. Lab Chip. 2012;12:3049–3054. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

