Cancer-mutation network and the number and specificity of driver mutations
- PMID: 29895694
- PMCID: PMC6042135
- DOI: 10.1073/pnas.1803155115
Cancer-mutation network and the number and specificity of driver mutations
Abstract
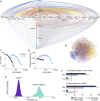
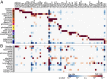
Cancer genomics has produced extensive information on cancer-associated genes, but the number and specificity of cancer-driver mutations remains a matter of debate. We constructed a bipartite network in which 7,665 tumors from 30 cancer types are connected via shared mutations in 198 previously identified cancer genes. We show that about 27% of the tumors can be assigned to statistically supported modules, most of which encompass one or two cancer types. The rest of the tumors belong to a diffuse network component suggesting lower gene specificity of driver mutations. Linear regression of the mutational loads in cancer genes was used to estimate the number of drivers required for the onset of different cancers. The mean number of drivers in known cancer genes is approximately two, with a range of one to five. Cancers that are associated with modules had more drivers than those from the diffuse network component, suggesting that unidentified and/or interchangeable drivers exist in the latter.
Keywords: bipartite networks; cancer types; community detection; driver mutations; passenger mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

