Tissue Specificity and Dynamics of Sex-Biased Gene Expression in a Common Frog Population with Differentiated, Yet Homomorphic, Sex Chromosomes
- PMID: 29895802
- PMCID: PMC6027210
- DOI: 10.3390/genes9060294
Tissue Specificity and Dynamics of Sex-Biased Gene Expression in a Common Frog Population with Differentiated, Yet Homomorphic, Sex Chromosomes
Abstract
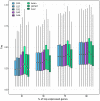
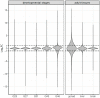
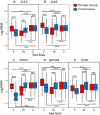
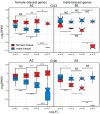
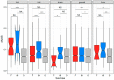
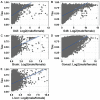
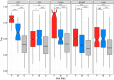
Sex-biased genes are central to the study of sexual selection, sexual antagonism, and sex chromosome evolution. We describe a comprehensive de novo assembled transcriptome in the common frog Rana temporaria based on five developmental stages and three adult tissues from both sexes, obtained from a population with karyotypically homomorphic but genetically differentiated sex chromosomes. This allows the study of sex-biased gene expression throughout development, and its effect on the rate of gene evolution while accounting for pleiotropic expression, which is known to negatively correlate with the evolutionary rate. Overall, sex-biased genes had little overlap among developmental stages and adult tissues. Late developmental stages and gonad tissues had the highest numbers of stage- or tissue-specific genes. We find that pleiotropic gene expression is a better predictor than sex bias for the evolutionary rate of genes, though it often interacts with sex bias. Although genetically differentiated, the sex chromosomes were not enriched in sex-biased genes, possibly due to a very recent arrest of XY recombination. These results extend our understanding of the developmental dynamics, tissue specificity, and genomic localization of sex-biased genes.
Keywords: adult tissues; development; gene expression; pleiotropy; rate of evolution; sex bias; sex chromosomes; tissue specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evolutionary and developmental dynamics of sex-biased gene expression in common frogs with proto-Y chromosomes.Genome Biol. 2018 Oct 5;19(1):156. doi: 10.1186/s13059-018-1548-4. Genome Biol. 2018. PMID: 30290841 Free PMC article.
-
Transcriptome assemblies for studying sex-biased gene expression in the guppy, Poecilia reticulata.BMC Genomics. 2014 May 26;15(1):400. doi: 10.1186/1471-2164-15-400. BMC Genomics. 2014. PMID: 24886435 Free PMC article.
-
Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution.Proc Natl Acad Sci U S A. 2013 Apr 16;110(16):6453-8. doi: 10.1073/pnas.1217027110. Epub 2013 Apr 1. Proc Natl Acad Sci U S A. 2013. PMID: 23547111 Free PMC article.
-
Sex-Biased Gene Expression.Annu Rev Genet. 2016 Nov 23;50:29-44. doi: 10.1146/annurev-genet-120215-035429. Epub 2016 Aug 26. Annu Rev Genet. 2016. PMID: 27574843 Review.
-
Ecology and the evolution of sex chromosomes.J Evol Biol. 2022 Dec;35(12):1601-1618. doi: 10.1111/jeb.14074. Epub 2022 Aug 11. J Evol Biol. 2022. PMID: 35950939 Review.
Cited by
-
A single QTL with large effect is associated with female functional virginity in an asexual parasitoid wasp.Mol Ecol. 2021 May;30(9):1979-1992. doi: 10.1111/mec.15863. Epub 2021 Mar 15. Mol Ecol. 2021. PMID: 33638236 Free PMC article.
-
The Diversity and Evolution of Sex Chromosomes in Frogs.Genes (Basel). 2021 Mar 26;12(4):483. doi: 10.3390/genes12040483. Genes (Basel). 2021. PMID: 33810524 Free PMC article. Review.
-
A bird-like genome from a frog: Mechanisms of genome size reduction in the ornate burrowing frog, Platyplectrum ornatum.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2011649118. doi: 10.1073/pnas.2011649118. Proc Natl Acad Sci U S A. 2021. PMID: 33836564 Free PMC article.
-
Hybridization and introgression between toads with different sex chromosome systems.Evol Lett. 2020 Aug 19;4(5):444-456. doi: 10.1002/evl3.191. eCollection 2020 Oct. Evol Lett. 2020. PMID: 33014420 Free PMC article.
-
Differential Gene Expression between Fungal Mating Types Is Associated with Sequence Degeneration.Genome Biol Evol. 2020 Apr 1;12(4):243-258. doi: 10.1093/gbe/evaa028. Genome Biol Evol. 2020. PMID: 32058544 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

