Comprehensive analysis of single molecule sequencing-derived complete genome and whole transcriptome of Hyposidra talaca nuclear polyhedrosis virus
- PMID: 29895987
- PMCID: PMC5997678
- DOI: 10.1038/s41598-018-27084-y
Comprehensive analysis of single molecule sequencing-derived complete genome and whole transcriptome of Hyposidra talaca nuclear polyhedrosis virus
Erratum in
-
Author Correction: Comprehensive analysis of single molecule sequencing-derived complete genome and whole transcriptome of Hyposidra talaca nuclear polyhedrosis virus.Sci Rep. 2018 Aug 31;8(1):13274. doi: 10.1038/s41598-018-31454-x. Sci Rep. 2018. PMID: 30171192 Free PMC article.
Abstract
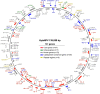
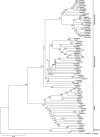
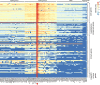
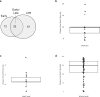
We sequenced the Hyposidra talaca NPV (HytaNPV) double stranded circular DNA genome using PacBio single molecule sequencing technology. We found that the HytaNPV genome is 139,089 bp long with a GC content of 39.6%. It encodes 141 open reading frames (ORFs) including the 37 baculovirus core genes, 25 genes conserved among lepidopteran baculoviruses, 72 genes known in baculovirus, and 7 genes unique to the HytaNPV genome. It is a group II alphabaculovirus that codes for the F protein and lacks the gp64 gene found in group I alphabaculovirus viruses. Using RNA-seq, we confirmed the expression of the ORFs identified in the HytaNPV genome. Phylogenetic analysis showed HytaNPV to be closest to BusuNPV, SujuNPV and EcobNPV that infect other tea pests, Buzura suppressaria, Sucra jujuba, and Ectropis oblique, respectively. We identified repeat elements and a conserved non-coding baculovirus element in the genome. Analysis of the putative promoter sequences identified motif consistent with the temporal expression of the genes observed in the RNA-seq data.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Patel VA, Shah SK. Tea Production in India: Challenges and Opportunities. Journal of Tea Science Research. 2016;6:1–6.
-
- Rabindra RJ. Sustinable pest management in tea: Prospects and challenges. Two and a Bud. 2012;59:1–10.
-
- Sinu P, Mandal P, Antony B. Range expansion of Hyposidra talaca (Geometridae: Lepidoptera), a major pest, to Northeastern Indian tea plantations: Change of weather and anti-predatory behaviour of the pest as possible causes. International Journal of Tropical Insect Science. 2001;31:242–248. doi: 10.1017/S174275841100035X. - DOI
-
- Chutia BC, Rahman A, Sarmah M, Barthakur BK, Borthakur M. Hyposidra talaca (Walker): A major defoliating pest of teas in North East India. Two and a Bud. 2012;59:17–20.
-
- Sannigrahi S, Talukdar T. Pesticide use patterns in Dooars tea industry. Two Bud. 2003;50:35–38.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

