Analysis of Beijing Douzhir Microbiota by High-Throughput Sequencing and Isolation of Acidogenic, Starch-Flocculating Strains
- PMID: 29896188
- PMCID: PMC5987674
- DOI: 10.3389/fmicb.2018.01142
Analysis of Beijing Douzhir Microbiota by High-Throughput Sequencing and Isolation of Acidogenic, Starch-Flocculating Strains
Abstract
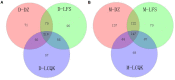
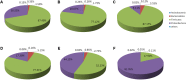
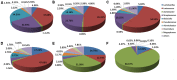
Beijing Douzhir is a traditional Chinese fermented drink produced by the natural fermentation of mung beans as the raw material. Ma tofu is an edible by-product of Douzhir processing. Douzhir microbiota, particularly bacteria involved in the natural fermentation process, has not been clearly established, resulting in limited industrial Douzhir production. Here, three uncooked Douzhir samples (D group) and three uncooked Ma tofu samples (M group) (two replicates per sample) were collected from three manufacturers in different locations in Beijing. The composition and diversity of the bacterial communities in each sample were analyzed by high-throughput sequencing. In total, 637 operational taxonomic units (OTUs) were revealed in the D group through database alignment, and 656 OTUs were found in the M group. The Chao, ACE, and Shannon indices were not significantly different in Douzhir samples from different manufacturers (p > 0.05). Representatives of six phyla were found in all 12 samples. Dominant bacteria were isolated and identified using mung bean juice as the growth medium. In both Douzhir and Ma tofu samples, dominant bacteria belonging to Firmicutes and Proteobacteria comprised > 94% of the total microbiota. The dominant bacteria included members of the Lactococcus, Acetobacter, Streptococcus, and Lactobacillus genera. Considering the dominant-microbiota information, we employed a plate-separation technique and isolated two strains of acid-producing bacteria from the Douzhir and Ma tofu samples with starch-flocculating activity: Acetobacter indonesiensis and Lactococcus lactis subsp. lactis. Such strains can serve as a foundation for the standardized industrial production of Douzhir.
Keywords: Douzhir; Ma tofu; acidogenic; high-throughput sequencing; isolation; starch flocculation.
Figures



Similar articles
-
[Analysis of the dynamic changes in gut microbiota in patients with extremely severe burns by 16S ribosomal RNA high-throughput sequencing technology].Zhonghua Shao Shang Za Zhi. 2020 Dec 20;36(12):1159-1166. doi: 10.3760/cma.j.cn501120-20200518-00271. Zhonghua Shao Shang Za Zhi. 2020. PMID: 33379852 Chinese.
-
Effects of temperature on paocai bacterial succession revealed by culture-dependent and culture-independent methods.Int J Food Microbiol. 2020 Mar 16;317:108463. doi: 10.1016/j.ijfoodmicro.2019.108463. Epub 2019 Nov 29. Int J Food Microbiol. 2020. PMID: 31809966
-
Analyses of physicochemical properties, bacterial microbiota, and lactic acid bacteria of fresh camel milk collected in Inner Mongolia.J Dairy Sci. 2020 Jan;103(1):106-116. doi: 10.3168/jds.2019-17023. Epub 2019 Oct 16. J Dairy Sci. 2020. PMID: 31629514
-
High-throughput sequencing and culture-based approaches to analyze microbial diversity associated with chemical changes in naturally fermented tofu whey, a traditional Chinese tofu-coagulant.Food Microbiol. 2018 Dec;76:69-77. doi: 10.1016/j.fm.2018.04.004. Epub 2018 Apr 11. Food Microbiol. 2018. PMID: 30166192
-
High-Throughput Sequence Analyses of Bacterial Communities and Multi-Mycotoxin Profiling During Processing of Different Formulations of Kunu, a Traditional Fermented Beverage.Front Microbiol. 2019 Jan 9;9:3282. doi: 10.3389/fmicb.2018.03282. eCollection 2018. Front Microbiol. 2019. PMID: 30687270 Free PMC article.
Cited by
-
The impact of lactic acid bacteria inoculation on the fermentation and metabolomic dynamics of indigenous Beijing douzhi microbial communities.Front Microbiol. 2024 Jul 30;15:1435834. doi: 10.3389/fmicb.2024.1435834. eCollection 2024. Front Microbiol. 2024. PMID: 39139380 Free PMC article.
-
Identification, Genome Characterization, and Growth Optimization of Paenibacillus peoriae MHJL1 for Biocontrol and Growth Promotion of Cotton Seedlings.Microorganisms. 2025 Jan 24;13(2):261. doi: 10.3390/microorganisms13020261. Microorganisms. 2025. PMID: 40005628 Free PMC article.
-
Fermentation improves flavors, bioactive substances, and antioxidant capacity of Bian-Que Triple-Bean Soup by lactic acid bacteria.Front Microbiol. 2023 Jul 18;14:1152654. doi: 10.3389/fmicb.2023.1152654. eCollection 2023. Front Microbiol. 2023. PMID: 37533834 Free PMC article.
-
Revealing the Characteristics and Correlations Among Microbial Communities, Functional Genes, and Vital Metabolites Through Metagenomics in Henan Mung Bean Sour.Microorganisms. 2025 Apr 7;13(4):845. doi: 10.3390/microorganisms13040845. Microorganisms. 2025. PMID: 40284681 Free PMC article.
-
Mung Bean Starch-Derived Fermented Liquid Alleviates Constipation via 5-HT Modulation and Gut Microbiota Regulation: An In Vivo Study.Foods. 2025 Jul 16;14(14):2483. doi: 10.3390/foods14142483. Foods. 2025. PMID: 40724304 Free PMC article.
References
-
- Chang Y. H., Lin C. L., Chen J. C. (2006). Characteristics of mung bean starch isolated by using lactic acid fermentation solution as the steeping liquor. Food Chem. 99 794–802. 10.1016/j.foodchem.2005.07.060 - DOI
-
- Chen Y. X., Cen L. J., Jiang T. M. (2013). Explore and fermentation test of fermented soya-bean milk (Beijing Douzhi) advantage bacterium group. Food Sci. Technol. 28 67–70.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

