Genomics of the Uncultivated, Periodontitis-Associated Bacterium Tannerella sp. BU045 (Oral Taxon 808)
- PMID: 29896567
- PMCID: PMC5989130
- DOI: 10.1128/mSystems.00018-18
Genomics of the Uncultivated, Periodontitis-Associated Bacterium Tannerella sp. BU045 (Oral Taxon 808)
Abstract
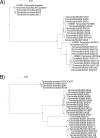
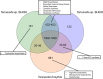
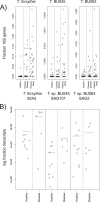
Despite decades of research into the human oral microbiome, many species remain uncultivated. The technique of single-cell whole-genome amplification and sequencing provides a means of deriving genome sequences for species that can be informative on biological function and suggest pathways to cultivation. Tannerella forsythia has long been known to be highly associated with chronic periodontitis and to cause periodontitis-like symptoms in experimental animals, and Tannerella sp. BU045 (human oral taxon 808) is an uncultivated relative of this organism. In this work, we extend our previous sequencing of the Tannerella sp. BU063 (human oral taxon 286) genome by sequencing amplified genomes from 11 cells of Tannerella sp. BU045, including 3 genomes that are at least 90% complete. Tannerella sp. BU045 is more closely related to Tannerella sp. BU063 than to T. forsythia by gene content and average nucleotide identity. However, two independent data sets of association with periodontitis, one based on 16S rRNA gene abundance and the other based on gene expression in a metatranscriptomic data set, show that Tannerella sp. BU045 is more highly associated with disease than Tannerella sp. BU063. Comparative genomics shows genes and functions that are shared or unique to the different species, which may direct further research of the pathogenesis of chronic periodontitis. IMPORTANCE Periodontitis (gum disease) affects 47% of adults over 30 in the United States (P. I. Eke, B. A. Dye, L. Wei, G. O. Thornton-Evans, R. J. Genco, et al., J Dent Res 91:914-920, 2012), and it cost between $39 and $396 billion worldwide in 2015 (A. J. Righolt, M. Jevdjevic, W. Marcenes, and S. Listl, J Dent Res, 17 January 2018, https://doi.org/10.1177/0022034517750572). Many bacteria associated with the disease are known only by the DNA sequence of their 16S rRNA gene. In this publication, amplification and sequencing of DNA from single bacterial cells are used to obtain nearly complete genomes of Tannerella sp. BU045, a species of bacteria that is more prevalent in patients with periodontitis than in healthy patients. Comparing the complete genome of this bacterium to genomes of related bacterial species will help to better understand periodontitis and may help to grow this organism in pure culture, which would allow a better understanding of its role in the mouth.
Keywords: Tannerella; WGA; oral microbiology; periodontitis; single cell.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

