DEPTOR at the Nexus of Cancer, Metabolism, and Immunity
- PMID: 29897294
- PMCID: PMC6335100
- DOI: 10.1152/physrev.00064.2017
DEPTOR at the Nexus of Cancer, Metabolism, and Immunity
Abstract
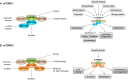
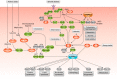
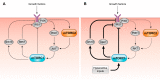
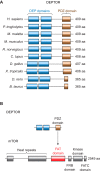
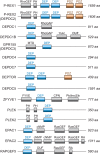
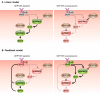
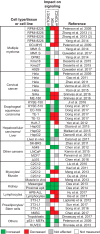
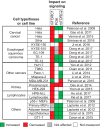
DEP domain-containing mechanistic target of rapamycin (mTOR)-interacting protein (DEPTOR) is an important modulator of mTOR, a kinase at the center of two important protein complexes named mTORC1 and mTORC2. These highly studied complexes play essential roles in regulating growth, metabolism, and immunity in response to mitogens, nutrients, and cytokines. Defects in mTOR signaling have been associated with the development of many diseases, including cancer and diabetes, and approaches aiming at modulating mTOR activity are envisioned as an attractive strategy to improve human health. DEPTOR interaction with mTOR represses its kinase activity and rewires the mTOR signaling pathway. Over the last years, several studies have revealed key roles for DEPTOR in numerous biological and pathological processes. Here, we provide the current state of the knowledge regarding the cellular and physiological functions of DEPTOR by focusing on its impact on the mTOR pathway and its role in promoting health and disease.
Figures
Similar articles
-
Regulation of human mTOR complexes by DEPTOR.Elife. 2021 Sep 14;10:e70871. doi: 10.7554/eLife.70871. Elife. 2021. PMID: 34519268 Free PMC article.
-
Deptor: not only a mTOR inhibitor.J Exp Clin Cancer Res. 2017 Jan 13;36(1):12. doi: 10.1186/s13046-016-0484-y. J Exp Clin Cancer Res. 2017. PMID: 28086984 Free PMC article. Review.
-
TGFβ-induced deptor suppression recruits mTORC1 and not mTORC2 to enhance collagen I (α2) gene expression.PLoS One. 2014 Oct 15;9(10):e109608. doi: 10.1371/journal.pone.0109608. eCollection 2014. PLoS One. 2014. PMID: 25333702 Free PMC article.
-
DEPTOR-mTOR Signaling Is Critical for Lipid Metabolism and Inflammation Homeostasis of Lymphocytes in Human PBMC Culture.J Immunol Res. 2017;2017:5252840. doi: 10.1155/2017/5252840. Epub 2017 Feb 27. J Immunol Res. 2017. PMID: 28349073 Free PMC article.
-
Research progress on the regulation mechanism of DEPTOR expression and its role in tumorigenesis.Neoplasma. 2025 Apr;72(1-2):1-15. doi: 10.4149/neo_2025_241203N504. Epub 2025 Mar 28. Neoplasma. 2025. PMID: 40162510 Review.
Cited by
-
Skeletal diseases caused by mutations in PTH1R show aberrant differentiation of skeletal progenitors due to dysregulation of DEPTOR.Front Cell Dev Biol. 2023 Jan 16;10:963389. doi: 10.3389/fcell.2022.963389. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36726589 Free PMC article.
-
DEPTOR modulates activation responses in CD4+ T cells and enhances immunoregulation following transplantation.Am J Transplant. 2019 Jan;19(1):77-88. doi: 10.1111/ajt.14995. Epub 2018 Aug 17. Am J Transplant. 2019. PMID: 29969188 Free PMC article.
-
The mTORC2 signaling network: targets and cross-talks.Biochem J. 2024 Jan 25;481(2):45-91. doi: 10.1042/BCJ20220325. Biochem J. 2024. PMID: 38270460 Free PMC article. Review.
-
DEPTOR stabilizes ErbB2 to promote the proliferation and survival of ErbB2-positive breast cancer cells.Theranostics. 2021 Apr 19;11(13):6355-6369. doi: 10.7150/thno.51286. eCollection 2021. Theranostics. 2021. PMID: 33995662 Free PMC article.
-
USF1-ATRAP-PBX3 Axis Promote Breast Cancer Glycolysis and Malignant Phenotype by Activating AKT/mTOR Signaling.Int J Biol Sci. 2022 Mar 14;18(6):2452-2471. doi: 10.7150/ijbs.69134. eCollection 2022. Int J Biol Sci. 2022. PMID: 35414770 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

