iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites
- PMID: 29897410
- PMCID: PMC6556904
- DOI: 10.1093/bib/bby028
iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites
Abstract
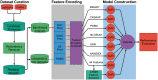
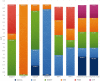
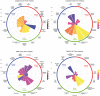
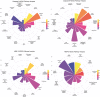
Regulation of proteolysis plays a critical role in a myriad of important cellular processes. The key to better understanding the mechanisms that control this process is to identify the specific substrates that each protease targets. To address this, we have developed iProt-Sub, a powerful bioinformatics tool for the accurate prediction of protease-specific substrates and their cleavage sites. Importantly, iProt-Sub represents a significantly advanced version of its successful predecessor, PROSPER. It provides optimized cleavage site prediction models with better prediction performance and coverage for more species-specific proteases (4 major protease families and 38 different proteases). iProt-Sub integrates heterogeneous sequence and structural features and uses a two-step feature selection procedure to further remove redundant and irrelevant features in an effort to improve the cleavage site prediction accuracy. Features used by iProt-Sub are encoded by 11 different sequence encoding schemes, including local amino acid sequence profile, secondary structure, solvent accessibility and native disorder, which will allow a more accurate representation of the protease specificity of approximately 38 proteases and training of the prediction models. Benchmarking experiments using cross-validation and independent tests showed that iProt-Sub is able to achieve a better performance than several existing generic tools. We anticipate that iProt-Sub will be a powerful tool for proteome-wide prediction of protease-specific substrates and their cleavage sites, and will facilitate hypothesis-driven functional interrogation of protease-specific substrate cleavage and proteolytic events.
Keywords: cleavage site; five-step rule; machine learning; protease; sequence analysis; substrate.
© The Author(s) 2018. Published by Oxford University Press.
Figures





Similar articles
-
PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites.PLoS One. 2012;7(11):e50300. doi: 10.1371/journal.pone.0050300. Epub 2012 Nov 29. PLoS One. 2012. PMID: 23209700 Free PMC article.
-
Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: a comprehensive revisit and benchmarking of existing methods.Brief Bioinform. 2019 Nov 27;20(6):2150-2166. doi: 10.1093/bib/bby077. Brief Bioinform. 2019. PMID: 30184176 Free PMC article. Review.
-
Procleave: Predicting Protease-specific Substrate Cleavage Sites by Combining Sequence and Structural Information.Genomics Proteomics Bioinformatics. 2020 Feb;18(1):52-64. doi: 10.1016/j.gpb.2019.08.002. Epub 2020 May 12. Genomics Proteomics Bioinformatics. 2020. PMID: 32413515 Free PMC article.
-
PROSPERous: high-throughput prediction of substrate cleavage sites for 90 proteases with improved accuracy.Bioinformatics. 2018 Feb 15;34(4):684-687. doi: 10.1093/bioinformatics/btx670. Bioinformatics. 2018. PMID: 29069280 Free PMC article.
-
Bioinformatic approaches for predicting substrates of proteases.J Bioinform Comput Biol. 2011 Feb;9(1):149-78. doi: 10.1142/s0219720011005288. J Bioinform Comput Biol. 2011. PMID: 21328711 Review.
Cited by
-
PD-L2 Is Constitutively Expressed in Normal and Malignant Urothelium.Front Oncol. 2021 Feb 25;11:626748. doi: 10.3389/fonc.2021.626748. eCollection 2021. Front Oncol. 2021. PMID: 33718196 Free PMC article.
-
Bastion3: a two-layer ensemble predictor of type III secreted effectors.Bioinformatics. 2019 Jun 1;35(12):2017-2028. doi: 10.1093/bioinformatics/bty914. Bioinformatics. 2019. PMID: 30388198 Free PMC article.
-
ADCdb: the database of antibody-drug conjugates.Nucleic Acids Res. 2024 Jan 5;52(D1):D1097-D1109. doi: 10.1093/nar/gkad831. Nucleic Acids Res. 2024. PMID: 37831118 Free PMC article.
-
Toward more accurate prediction of caspase cleavage sites: a comprehensive review of current methods, tools and features.Brief Bioinform. 2019 Sep 27;20(5):1669-1684. doi: 10.1093/bib/bby041. Brief Bioinform. 2019. PMID: 29860277 Free PMC article.
-
Mapping specificity, cleavage entropy, allosteric changes and substrates of blood proteases in a high-throughput screen.Nat Commun. 2021 Mar 16;12(1):1693. doi: 10.1038/s41467-021-21754-8. Nat Commun. 2021. PMID: 33727531 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

