Traditional and systems biology based drug discovery for the rare tumor syndrome neurofibromatosis type 2
- PMID: 29897904
- PMCID: PMC5999111
- DOI: 10.1371/journal.pone.0197350
Traditional and systems biology based drug discovery for the rare tumor syndrome neurofibromatosis type 2
Abstract
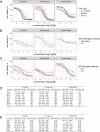
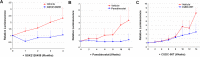
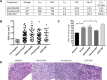
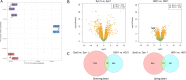
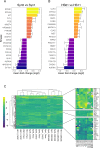
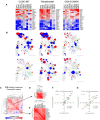
Neurofibromatosis 2 (NF2) is a rare tumor suppressor syndrome that manifests with multiple schwannomas and meningiomas. There are no effective drug therapies for these benign tumors and conventional therapies have limited efficacy. Various model systems have been created and several drug targets have been implicated in NF2-driven tumorigenesis based on known effects of the absence of merlin, the product of the NF2 gene. We tested priority compounds based on known biology with traditional dose-concentration studies in meningioma and schwann cell systems. Concurrently, we studied functional kinome and gene expression in these cells pre- and post-treatment to determine merlin deficient molecular phenotypes. Cell viability results showed that three agents (GSK2126458, Panobinostat, CUDC-907) had the greatest activity across schwannoma and meningioma cell systems, but merlin status did not significantly influence response. In vivo, drug effect was tumor specific with meningioma, but not schwannoma, showing response to GSK2126458 and Panobinostat. In culture, changes in both the transcriptome and kinome in response to treatment clustered predominantly based on tumor type. However, there were differences in both gene expression and functional kinome at baseline between meningioma and schwannoma cell systems that may form the basis for future selective therapies. This work has created an openly accessible resource (www.synapse.org/SynodosNF2) of fully characterized isogenic schwannoma and meningioma cell systems as well as a rich data source of kinome and transcriptome data from these assay systems before and after treatment that enables single and combination drug discovery based on molecular phenotype.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Evans DG. Neurofibromatosis type 2. Handb Clin Neurol. 2015;132: 87–96. doi: 10.1016/B978-0-444-62702-5.00005-6 - DOI - PubMed
-
- Hexter A, Jones A, Joe H, Heap L, Smith MJ, Wallace AJ, et al. Clinical and molecular predictors of mortality in neurofibromatosis 2: a UK national analysis of 1192 patients. J Med Genet. 2015;52: 699–705. doi: 10.1136/jmedgenet-2015-103290 - DOI - PubMed
-
- Domingues P, Gonzalez-Tablas M, Otero A, Pascual D, Ruiz L, Miranda D, et al. Genetic/molecular alterations of meningiomas and the signaling pathways targeted. Oncotarget. 2015;6: 10671–10688. doi: 10.18632/oncotarget.3870 - DOI - PMC - PubMed
-
- Mallory GW, Pollock BE, Foote RL, Carlson ML, Driscoll CL, Link MJ. Stereotactic radiosurgery for neurofibromatosis 2-associated vestibular schwannomas: toward dose optimization for tumor control and functional outcomes. Neurosurgery. 2014;74: 292–300; discussion 300–1. doi: 10.1227/NEU.0000000000000264 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

