BATF regulates the expression of Nfil3, Wnt10a and miR155hg for efficient induction of antibody class switch recombination in mice
- PMID: 29898247
- PMCID: PMC6357966
- DOI: 10.1002/eji.201747360
BATF regulates the expression of Nfil3, Wnt10a and miR155hg for efficient induction of antibody class switch recombination in mice
Abstract
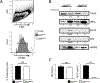
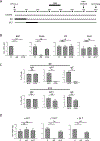
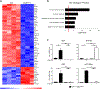
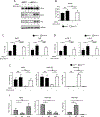
BATF functions in T cells and B cells to control the host response to antigen and promote the production of class switched immunoglobulins. In this study, we demonstrate that BATF expression increases rapidly, and transiently, following B cell stimulation and use an inducible murine model of BATF deletion to show that this induction is necessary, and sufficient, for immunoglobulin (Ig) class switch recombination (CSR). We examine two genes (Nfil3 and miR155gh) that are positively regulated, and one gene (Wnt10a) that is negatively regulated by BATF during CSR. These genes play essential roles in CSR and each impacts the expression and/or function of the others. Our observations allow these targets of BATF regulation to be positioned in a network upstream of the activation of germline transcripts (GLT) from the IgH locus and of transcriptional activation of Aicda - the gene encoding the enzyme directing Ig gene rearrangements. This work extends the knowledge of the molecular control of CSR and, importantly, positions the induction and function of BATF as an early event in this process.
Keywords: B cells; BATF; class switch recombination; signaling pathways; transcription regulation.
© 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
The authors declare no commercial or financial conflict of interest.
Figures

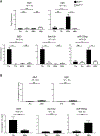
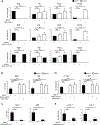

References
-
- Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013; 13:499–509. - PubMed
-
- Sopel N, Graser A, Mousset S, Finotto S. The transcription factor BATF modulates cytokine-mediated responses in T cells. Cytokine Growth Factor Rev. 2016; 30:39–45. - PubMed
-
- Dorsey M, Tae H, Sollenberger K, Mascarenhas N, Johansen L, Taparowsky E. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 1995; 11:2255–2265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

