Autophagy Differentially Regulates Insulin Production and Insulin Sensitivity
- PMID: 29898399
- PMCID: PMC6054876
- DOI: 10.1016/j.celrep.2018.05.032
Autophagy Differentially Regulates Insulin Production and Insulin Sensitivity
Abstract
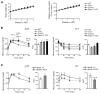
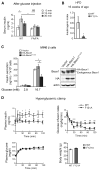
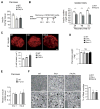
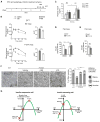
Autophagy, a stress-induced lysosomal degradative pathway, has been assumed to exert similar metabolic effects in different organs. Here, we establish a model where autophagy plays different roles in insulin-producing β cells versus insulin-responsive cells, utilizing knockin (Becn1F121A) mice manifesting constitutively active autophagy. With a high-fat-diet challenge, the autophagy-hyperactive mice unexpectedly show impaired glucose tolerance, but improved insulin sensitivity, compared to mice with normal autophagy. Autophagy hyperactivation enhances insulin signaling, via suppressing ER stress in insulin-responsive cells, but decreases insulin secretion by selectively sequestrating and degrading insulin granule vesicles in β cells, a process we term "vesicophagy." The reduction in insulin storage, insulin secretion, and glucose tolerance is reversed by transient treatment of autophagy inhibitors. Thus, β cells and insulin-responsive tissues require different autophagy levels for optimal function. To improve insulin sensitivity without hampering secretion, acute or intermittent, rather than chronic, activation of autophagy should be considered in diabetic therapy development.
Keywords: Becn1; autophagosome; autophagy; glucose tolerance; insulin granule; insulin sensitivity; insulin-responsive tissue; type 2 diabetes; vesicophagy; β cell.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
The BECN1-BCL2 complex regulates insulin secretion and storage in mice.Autophagy. 2018;14(11):2026-2028. doi: 10.1080/15548627.2018.1502566. Epub 2018 Aug 21. Autophagy. 2018. PMID: 30081744 Free PMC article.
References
-
- Andersson A, Olsson S, Tjälve H. Chloroquine inhibits the insulin production of isolated pancreatic islets. Biochem Pharmacol. 1980;29:1729–1735. - PubMed
-
- Chatterjee AK, Schatz H. Effect of chloroquine on biosynthesis, release and degradation of insulin in isolated islets of rat pancreas. Diabetes Res Clin Pract. 1988;5:9–15. - PubMed
-
- Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, Lakey ND, Culpepper J, Moore KJ, Breitbart RE, et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. - PubMed
-
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

