Integrated Molecular Characterization of Testicular Germ Cell Tumors
- PMID: 29898407
- PMCID: PMC6075738
- DOI: 10.1016/j.celrep.2018.05.039
Integrated Molecular Characterization of Testicular Germ Cell Tumors
Abstract
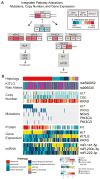
We studied 137 primary testicular germ cell tumors (TGCTs) using high-dimensional assays of genomic, epigenomic, transcriptomic, and proteomic features. These tumors exhibited high aneuploidy and a paucity of somatic mutations. Somatic mutation of only three genes achieved significance-KIT, KRAS, and NRAS-exclusively in samples with seminoma components. Integrated analyses identified distinct molecular patterns that characterized the major recognized histologic subtypes of TGCT: seminoma, embryonal carcinoma, yolk sac tumor, and teratoma. Striking differences in global DNA methylation and microRNA expression between histology subtypes highlight a likely role of epigenomic processes in determining histologic fates in TGCTs. We also identified a subset of pure seminomas defined by KIT mutations, increased immune infiltration, globally demethylated DNA, and decreased KRAS copy number. We report potential biomarkers for risk stratification, such as miRNA specifically expressed in teratoma, and others with molecular diagnostic potential, such as CpH (CpA/CpC/CpT) methylation identifying embryonal carcinomas.
Keywords: DNA methylation; KIT; The Cancer Genome Atlas; copy number; exome sequencing; miR-375; nonseminoma; seminoma; testicular germ cell tumors.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
A.C. and M.M. receive research funding from Bayer AG. J.M.S. is a founder and part owner of Five3 Genomics LLC and a member of its scientific advisory board, which owns the license to PARADIGM software.
Figures






References
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. - PMC - PubMed
-
- Banks K, Tuazon E, Berhane K, Koh CJ, De Filippo RE, Chang A, Kim SS, Daneshmand S, Davis-Dao C, Lewinger JP, et al. Cryptorchidism and testicular germ cell tumors: comprehensive meta-analysis reveals that association between these conditions diminished over time and is modified by clinical characteristics. Front Endocrinol (Lausanne) 2013;3:182. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32 GM008638/GM/NIGMS NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- KL2 TR001879/TR/NCATS NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R01 CA180778/CA/NCI NIH HHS/United States
- U24 CA210949/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- U24 CA210969/CA/NCI NIH HHS/United States
- U24 CA210988/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

