Replication Fork Breakage and Restart in Escherichia coli
- PMID: 29898897
- PMCID: PMC6094043
- DOI: 10.1128/MMBR.00013-18
Replication Fork Breakage and Restart in Escherichia coli
Abstract
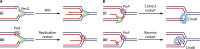
In all organisms, replication impairments are an important source of genome rearrangements, mainly because of the formation of double-stranded DNA (dsDNA) ends at inactivated replication forks. Three reactions for the formation of dsDNA ends at replication forks were originally described for Escherichia coli and became seminal models for all organisms: the encounter of replication forks with preexisting single-stranded DNA (ssDNA) interruptions, replication fork reversal, and head-to-tail collisions of successive replication rounds. Here, we first review the experimental evidence that now allows us to know when, where, and how these three different reactions occur in E. coli. Next, we recall our recent studies showing that in wild-type E. coli, spontaneous replication fork breakage occurs in 18% of cells at each generation. We propose that it results from the replication of preexisting nicks or gaps, since it does not involve replication fork reversal or head-to-tail fork collisions. In the recB mutant, deficient for double-strand break (DSB) repair, fork breakage triggers DSBs in the chromosome terminus during cell division, a reaction that is heritable for several generations. Finally, we recapitulate several observations suggesting that restart from intact inactivated replication forks and restart from recombination intermediates require different sets of enzymatic activities. The finding that 18% of cells suffer replication fork breakage suggests that DNA remains intact at most inactivated forks. Similarly, only 18% of cells need the helicase loader for replication restart, which leads us to speculate that the replicative helicase remains on DNA at intact inactivated replication forks and is reactivated by the replication restart proteins.
Keywords: PriA; RecA; RecBC; RecBCD; RecG; RuvAB; chromosome terminus; double-strand break; recombination; replication fork reversal; replication restart.
Copyright © 2018 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

