Recapitulating muscle disease phenotypes with myotonic dystrophy 1 induced pluripotent stem cells: a tool for disease modeling and drug discovery
- PMID: 29898953
- PMCID: PMC6078411
- DOI: 10.1242/dmm.034728
Recapitulating muscle disease phenotypes with myotonic dystrophy 1 induced pluripotent stem cells: a tool for disease modeling and drug discovery
Abstract
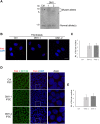
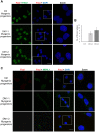
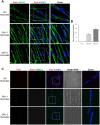
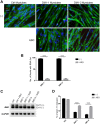
Myotonic dystrophy 1 (DM1) is a multisystem disorder primarily affecting the central nervous system, heart and skeletal muscle. It is caused by an expansion of the CTG trinucleotide repeats in the 3' untranslated region of the DMPK gene. Although patient myoblasts have been used for studying the disease in vitro, the invasiveness as well as the low accessibility to muscle biopsies motivate the development of alternative reliable myogenic models. Here, we established two DM1 induced pluripotent stem (iPS) cell lines from patient-derived fibroblasts and, using the PAX7 conditional expression system, differentiated these into myogenic progenitors and, subsequently, terminally differentiated myotubes. Both DM1 myogenic progenitors and myotubes were found to express the intranuclear RNA foci exhibiting sequestration of MBNL1. Moreover, we found the DM1-related mis-splicing, namely BIN1 exon 11 in DM1 myotubes. We used this model to test a specific therapy, antisense oligonucleotide treatment, and found that this efficiently abolished RNA foci and rescued BIN1 mis-splicing in DM1 iPS cell-derived myotubes. Together, our results demonstrate that myotubes derived from DM1 iPS cells recapitulate the critical molecular features of DM1 and are sensitive to antisense oligonucleotide treatment, confirming that these cells can be used for in vitro disease modeling and candidate drug testing or screening.This article has an associated First Person interview with the first author of the paper.
Keywords: Induced pluripotent stem (iPS) cells; Muscular dystrophy; Myotonic dystrophy; PAX7; RNA foci; Skeletal myogenesis.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
References
-
- Arandel L., Polay Espinoza M., Matloka M., Bazinet A., De Dea Diniz D., Naouar N., Rau F., Jollet A., Edom-Vovard F., Mamchaoui K. et al. (2017). Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis. Model Mech. 10, 487-497. 10.1242/dmm.027367 - DOI - PMC - PubMed
-
- Chen I.-P., Fukuda K., Fusaki N., Iida A., Hasegawa M., Lichtler A. and Reichenberger E. J. (2013). Induced pluripotent stem cell reprogramming by integration-free Sendai virus vectors from peripheral blood of patients with craniometaphyseal dysplasia. Cell Reprogram 15, 503-513. 10.1089/cell.2013.0037 - DOI - PMC - PubMed
-
- Darabi R., Arpke R. W., Irion S., Dimos J. T., Grskovic M., Kyba M. and Perlingeiro R. C. R. (2012). Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10, 610-619. 10.1016/j.stem.2012.02.015 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

