Expression of a Structural Protein of the Mycovirus FgV-ch9 Negatively Affects the Transcript Level of a Novel Symptom Alleviation Factor and Causes Virus Infection-Like Symptoms in Fusarium graminearum
- PMID: 29899100
- PMCID: PMC6096815
- DOI: 10.1128/JVI.00326-18
Expression of a Structural Protein of the Mycovirus FgV-ch9 Negatively Affects the Transcript Level of a Novel Symptom Alleviation Factor and Causes Virus Infection-Like Symptoms in Fusarium graminearum
Abstract
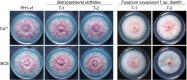
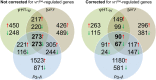
Infections of fungi by mycoviruses are often symptomless but sometimes also fatal, as they perturb sporulation, growth, and, if applicable, virulence of the fungal host. Hypovirulence-inducing mycoviruses, therefore, represent a powerful means to defeat fungal epidemics on crop plants. Infection with Fusarium graminearum virus China 9 (FgV-ch9), a double-stranded RNA (dsRNA) chrysovirus-like mycovirus, debilitates Fusarium graminearum, the causal agent of fusarium head blight. In search for potential symptom alleviation or aggravation factors in F. graminearum, we consecutively infected a custom-made F. graminearum mutant collection with FgV-ch9 and found a mutant with constantly elevated expression of a gene coding for a putative mRNA-binding protein that did not show any disease symptoms despite harboring large amounts of virus. Deletion of this gene, named virus response 1 (vr1), resulted in phenotypes identical to those observed in the virus-infected wild type with respect to growth, reproduction, and virulence. Similarly, the viral structural protein coded on segment 3 (P3) caused virus infection-like symptoms when expressed in the wild type but not in the vr1 overexpression mutant. Gene expression analysis revealed a drastic downregulation of vr1 in the presence of virus and in mutants expressing P3. We conclude that symptom development and severity correlate with gene expression levels of vr1 This was confirmed by comparative transcriptome analysis, showing a large transcriptional overlap between the virus-infected wild type, the vr1 deletion mutant, and the P3-expressing mutant. Hence, vr1 represents a fundamental host factor for the expression of virus-related symptoms and helps us understand the underlying mechanism of hypovirulence.IMPORTANCE Virus infections of phytopathogenic fungi occasionally impair growth, reproduction, and virulence, a phenomenon referred to as hypovirulence. Hypovirulence-inducing mycoviruses, therefore, represent a powerful means to defeat fungal epidemics on crop plants. However, the poor understanding of the molecular basis of hypovirulence induction limits their application. Using the devastating fungal pathogen on cereal crops, Fusarium graminearum, we identified an mRNA binding protein (named virus response 1, vr1) which is involved in symptom expression. Downregulation of vr1 in the virus-infected fungus and vr1 deletion evoke virus infection-like symptoms, while constitutive expression overrules the cytopathic effects of the virus infection. Intriguingly, the presence of a specific viral structural protein is sufficient to trigger the fungal response, i.e., vr1 downregulation, and symptom development similar to virus infection. The advancements in understanding fungal infection and response may aid biological pest control approaches using mycoviruses or viral proteins to prevent future Fusarium epidemics.
Keywords: Chrysoviridae; FgV-ch9; Fusarium graminearum; fungal response; hypovirulence; mycovirus.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
Effects of the deletion and over-expression of Fusarium graminearum gene FgHal2 on host response to mycovirus Fusarium graminearum virus 1.Mol Plant Pathol. 2015 Sep;16(7):641-52. doi: 10.1111/mpp.12221. Epub 2015 Jan 29. Mol Plant Pathol. 2015. PMID: 25431083 Free PMC article.
-
Exploration of the interactions between mycoviruses and Fusarium graminearum.Adv Virus Res. 2020;106:123-144. doi: 10.1016/bs.aivir.2020.01.004. Epub 2020 Feb 5. Adv Virus Res. 2020. PMID: 32327146 Review.
-
Mycoviruses in Fusarium Species: An Update.Front Cell Infect Microbiol. 2019 Jul 18;9:257. doi: 10.3389/fcimb.2019.00257. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380300 Free PMC article. Review.
-
A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses.PLoS One. 2014 Jun 25;9(6):e100989. doi: 10.1371/journal.pone.0100989. eCollection 2014. PLoS One. 2014. PMID: 24964178 Free PMC article.
-
Duplications in the 3' termini of three segments of Fusarium graminearum virus China 9.Arch Virol. 2017 Mar;162(3):897-900. doi: 10.1007/s00705-016-3174-3. Epub 2016 Nov 25. Arch Virol. 2017. PMID: 27888409
Cited by
-
Stable overexpression and targeted gene deletion of the causative agent of ash dieback Hymenoscyphus fraxineus.Fungal Biol Biotechnol. 2023 Jan 13;10(1):1. doi: 10.1186/s40694-023-00149-y. Fungal Biol Biotechnol. 2023. PMID: 36639657 Free PMC article.
-
A Phenome-Wide Association Study of the Effects of Fusarium graminearum Transcription Factors on Fusarium Graminearum Virus 1 Infection.Front Microbiol. 2021 Feb 11;12:622261. doi: 10.3389/fmicb.2021.622261. eCollection 2021. Front Microbiol. 2021. PMID: 33643250 Free PMC article.
-
Mycologists and Virologists Align: Proposing Botrytis cinerea for Global Mycovirus Studies.Viruses. 2024 Sep 18;16(9):1483. doi: 10.3390/v16091483. Viruses. 2024. PMID: 39339959 Free PMC article. Review.
-
Chrysoviruses in Magnaporthe oryzae.Viruses. 2018 Dec 8;10(12):697. doi: 10.3390/v10120697. Viruses. 2018. PMID: 30544784 Free PMC article. Review.
-
Spray-induced gene silencing for crop protection: recent advances and emerging trends.Front Plant Sci. 2025 Feb 20;16:1527944. doi: 10.3389/fpls.2025.1527944. eCollection 2025. Front Plant Sci. 2025. PMID: 40051878 Free PMC article. Review.
References
-
- Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N. 2015. 50-Plus years of fungal viruses. Virology 479–480:356–368. - PubMed
-
- Buck KW. 1998. Molecular variability of viruses of fungi, p 53–72. In Bridge P, Couteaudier Y, Clarkson J (ed), Molecular variability of fungal pathogens. Springer Science & Business Media, Berlin, Germany.
-
- Nuss D. 2011. Mycoviruses, p 145–152. In Borkovich K, Ebbole D (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

