A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase
- PMID: 29899144
- PMCID: PMC6042107
- DOI: 10.1073/pnas.1806774115
A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase
Abstract
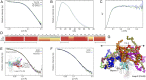
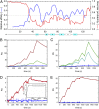
The intravascular processing of triglyceride-rich lipoproteins depends on lipoprotein lipase (LPL) and GPIHBP1, a membrane protein of endothelial cells that binds LPL within the subendothelial spaces and shuttles it to the capillary lumen. In the absence of GPIHBP1, LPL remains mislocalized within the subendothelial spaces, causing severe hypertriglyceridemia (chylomicronemia). The N-terminal domain of GPIHBP1, an intrinsically disordered region (IDR) rich in acidic residues, is important for stabilizing LPL's catalytic domain against spontaneous and ANGPTL4-catalyzed unfolding. Here, we define several important properties of GPIHBP1's IDR. First, a conserved tyrosine in the middle of the IDR is posttranslationally modified by O-sulfation; this modification increases both the affinity of GPIHBP1-LPL interactions and the ability of GPIHBP1 to protect LPL against ANGPTL4-catalyzed unfolding. Second, the acidic IDR of GPIHBP1 increases the probability of a GPIHBP1-LPL encounter via electrostatic steering, increasing the association rate constant (kon) for LPL binding by >250-fold. Third, we show that LPL accumulates near capillary endothelial cells even in the absence of GPIHBP1. In wild-type mice, we expect that the accumulation of LPL in close proximity to capillaries would increase interactions with GPIHBP1. Fourth, we found that GPIHBP1's IDR is not a key factor in the pathogenicity of chylomicronemia in patients with the GPIHBP1 autoimmune syndrome. Finally, based on biophysical studies, we propose that the negatively charged IDR of GPIHBP1 traverses a vast space, facilitating capture of LPL by capillary endothelial cells and simultaneously contributing to GPIHBP1's ability to preserve LPL structure and activity.
Keywords: autoimmune disease; electrostatic steering; hypertriglyceridemia; intravascular lipolysis; intrinsically disordered region.
Conflict of interest statement
Conflict of interest statement: S.G.Y. and Jay D. Horton were both principal investigators on a consortium grant. They did not collaborate directly.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

