Antithetic proportional-integral feedback for reduced variance and improved control performance of stochastic reaction networks
- PMID: 29899158
- PMCID: PMC6030643
- DOI: 10.1098/rsif.2018.0079
Antithetic proportional-integral feedback for reduced variance and improved control performance of stochastic reaction networks
Abstract
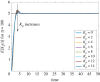
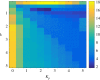
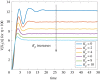
The ability of a cell to regulate and adapt its internal state in response to unpredictable environmental changes is called homeostasis and this ability is crucial for the cell's survival and proper functioning. Understanding how cells can achieve homeostasis, despite the intrinsic noise or randomness in their dynamics, is fundamentally important for both systems and synthetic biology. In this context, a significant development is the proposed antithetic integral feedback (AIF) motif, which is found in natural systems, and is known to ensure robust perfect adaptation for the mean dynamics of a given molecular species involved in a complex stochastic biomolecular reaction network. From the standpoint of applications, one drawback of this motif is that it often leads to an increased cell-to-cell heterogeneity or variance when compared to a constitutive (i.e. open-loop) control strategy. Our goal in this paper is to show that this performance deterioration can be countered by combining the AIF motif and a negative feedback strategy. Using a tailored moment closure method, we derive approximate expressions for the stationary variance for the controlled network that demonstrate that increasing the strength of the negative feedback can indeed decrease the variance, sometimes even below its constitutive level. Numerical results verify the accuracy of these results and we illustrate them by considering three biomolecular networks with two types of negative feedback strategies. Our computational analysis indicates that there is a trade-off between the speed of the settling-time of the mean trajectories and the stationary variance of the controlled species; i.e. smaller variance is associated with larger settling-time.
Keywords: antithetic integral control; cybergenetics; homeostasis; stochastic reaction networks.
© 2018 The Author(s).
Conflict of interest statement
We declare we have no competing interests.
Figures
















References
-
- CubaSamaniego C, Franco E. 2017. An ultrasensitive biomolecular network for robust feedback control. In 20th IFAC World Congress, Toulouse, France, pp. 11 437–11 443. Laxenburg, Austria: IFAC.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

