Deciphering the Adaptation of Corynebacterium glutamicum in Transition from Aerobiosis via Microaerobiosis to Anaerobiosis
- PMID: 29899275
- PMCID: PMC6027265
- DOI: 10.3390/genes9060297
Deciphering the Adaptation of Corynebacterium glutamicum in Transition from Aerobiosis via Microaerobiosis to Anaerobiosis
Abstract
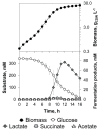
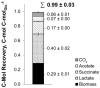
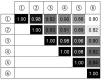
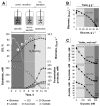
Zero-growth processes are a promising strategy for the production of reduced molecules and depict a steady transition from aerobic to anaerobic conditions. To investigate the adaptation of Corynebacterium glutamicum to altering oxygen availabilities, we conceived a triple-phase fermentation process that describes a gradual reduction of dissolved oxygen with a shift from aerobiosis via microaerobiosis to anaerobiosis. The distinct process phases were clearly bordered by the bacteria’s physiologic response such as reduced growth rate, biomass substrate yield and altered yield of fermentation products. During the process, sequential samples were drawn at six points and analyzed via RNA-sequencing, for metabolite concentrations and for enzyme activities. We found transcriptional alterations of almost 50% (1421 genes) of the entire protein coding genes and observed an upregulation of fermentative pathways, a rearrangement of respiration, and mitigation of the basic cellular mechanisms such as transcription, translation and replication as a transient response related to the installed oxygen dependent process phases. To investigate the regulatory regime, 18 transcriptionally altered (putative) transcriptional regulators were deleted, but none of the deletion strains showed noticeable growth kinetics under an oxygen restricted environment. However, the described transcriptional adaptation of C. glutamicum resolved to varying oxygen availabilities provides a useful basis for future process and strain engineering.
Keywords: Corynebacterium glutamicum; aerobiosis; anaerobiosis; microaerobiosis; transcriptional response; triple-phase process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Lactate production as representative of the fermentation potential of Corynebacterium glutamicum 2262 in a one-step process.Biosci Biotechnol Biochem. 2014;78(2):343-9. doi: 10.1080/09168451.2014.878219. Epub 2014 Apr 14. Biosci Biotechnol Biochem. 2014. PMID: 25036691
-
Physiological characteristics of Corynebacterium glutamicum as a cell factory under anaerobic conditions.Appl Microbiol Biotechnol. 2021 Aug;105(16-17):6173-6181. doi: 10.1007/s00253-021-11474-w. Epub 2021 Aug 17. Appl Microbiol Biotechnol. 2021. PMID: 34402937 Review.
-
Zero-growth bioprocesses: A challenge for microbial production strains and bioprocess engineering.Eng Life Sci. 2016 Nov 11;17(1):27-35. doi: 10.1002/elsc.201600108. eCollection 2017 Jan. Eng Life Sci. 2016. PMID: 32624726 Free PMC article. Review.
-
Corynebacterium glutamicum CgynfM encodes a dicarboxylate transporter applicable to succinate production.J Biosci Bioeng. 2019 Apr;127(4):465-471. doi: 10.1016/j.jbiosc.2018.10.004. Epub 2018 Nov 2. J Biosci Bioeng. 2019. PMID: 30392965
-
Anaerobic growth of Corynebacterium glutamicum via mixed-acid fermentation.Appl Environ Microbiol. 2015 Nov;81(21):7496-508. doi: 10.1128/AEM.02413-15. Epub 2015 Aug 14. Appl Environ Microbiol. 2015. PMID: 26276118 Free PMC article.
Cited by
-
Tailoring Corynebacterium glutamicum for Sustainable Biomanufacturing: From Traditional to Cutting-Edge Technologies.Mol Biotechnol. 2025 Jun 10. doi: 10.1007/s12033-025-01447-z. Online ahead of print. Mol Biotechnol. 2025. PMID: 40493161 Review.
-
Development of a workflow for the selection, identification and optimization of lactic acid bacteria with high γ-aminobutyric acid production.Sci Rep. 2023 Aug 22;13(1):13663. doi: 10.1038/s41598-023-40808-z. Sci Rep. 2023. PMID: 37608211 Free PMC article.
-
Physiological Response of Corynebacterium glutamicum to Indole.Microorganisms. 2020 Dec 8;8(12):1945. doi: 10.3390/microorganisms8121945. Microorganisms. 2020. PMID: 33302489 Free PMC article.
-
Genomics Characterization of an Engineered Corynebacterium glutamicum in Bioreactor Cultivation Under Ionic Liquid Stress.Front Bioeng Biotechnol. 2021 Nov 18;9:766674. doi: 10.3389/fbioe.2021.766674. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34869279 Free PMC article.
-
Corynebacterium glutamicum, a natural overproducer of succinic acid?Eng Life Sci. 2020 Jan 20;20(5-6):205-215. doi: 10.1002/elsc.201900141. eCollection 2020 Apr. Eng Life Sci. 2020. PMID: 32874184 Free PMC article.
References
-
- Ajinomoto Co., Inc. FY2015 Market and Other Information. [(accessed on 7 September 2017)]; Available online: https://www.ajinomoto.com/en/ir/event/presentation/main/09/teaserItems1/....
-
- Liebl W. Corynebacterium taxonomy. In: Eggeling L., Bott M., editors. Handbook of Corynebacterium Glutamicum. CRC Press; Boca Raton, FL, USA: 2005. pp. 9–34.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

