Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii
- PMID: 29899345
- PMCID: PMC5997985
- DOI: 10.1038/s41598-018-26689-7
Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii
Abstract
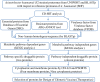
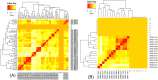
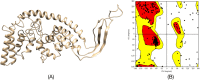
The emergence of drug-resistant Acinetobacter baumannii is the global health problem associated with high mortality and morbidity. Therefore it is high time to find a suitable therapeutics for this pathogen. In the present study, subtractive proteomics along with reverse vaccinology approaches were used to predict suitable therapeutics against A. baumannii. Using subtractive proteomics, we have identified promiscuous antigenic membrane proteins that contain the virulence factors, resistance factors and essentiality factor for this pathogenic bacteria. Selected promiscuous targeted membrane proteins were used for the design of chimeric-subunit vaccine with the help of reverse vaccinology. Available best tools and servers were used for the identification of MHC class I, II and B cell epitopes. All selected epitopes were further shortlisted computationally to know their immunogenicity, antigenicity, allergenicity, conservancy and toxicity potentials. Immunogenic predicted promiscuous peptides used for the development of chimeric subunit vaccine with immune-modulating adjuvants, linkers, and PADRE (Pan HLA-DR epitopes) amino acid sequence. Designed vaccine construct V4 also interact with the MHC, and TLR4/MD2 complex as confirm by docking and molecular dynamics simulation studies. Therefore designed vaccine construct V4 can be developed to control the host-pathogen interaction or infection caused by A. baumannii.
Conflict of interest statement
The authors declare no competing interests.
Figures





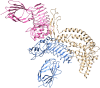


References
-
- Tiwari V, Rajeswari MR. Effect of Iron Availability on the Survival of Carbapenem-Resistant Acinetobacter baumannii: a Proteomic Approach. Journal of Proteomics & Bioinformatics. 2013;06:125–131. doi: 10.4172/jpb.1000270. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

