Network biology discovers pathogen contact points in host protein-protein interactomes
- PMID: 29899369
- PMCID: PMC5998135
- DOI: 10.1038/s41467-018-04632-8
Network biology discovers pathogen contact points in host protein-protein interactomes
Abstract
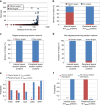
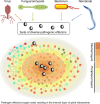
In all organisms, major biological processes are controlled by complex protein-protein interactions networks (interactomes), yet their structural complexity presents major analytical challenges. Here, we integrate a compendium of over 4300 phenotypes with Arabidopsis interactome (AI-1MAIN). We show that nodes with high connectivity and betweenness are enriched and depleted in conditional and essential phenotypes, respectively. Such nodes are located in the innermost layers of AI-1MAIN and are preferential targets of pathogen effectors. We extend these network-centric analyses to Cell Surface Interactome (CSILRR) and predict its 35 most influential nodes. To determine their biological relevance, we show that these proteins physically interact with pathogen effectors and modulate plant immunity. Overall, our findings contrast with centrality-lethality rule, discover fast information spreading nodes, and highlight the structural properties of pathogen targets in two different interactomes. Finally, this theoretical framework could possibly be applicable to other inter-species interactomes to reveal pathogen contact points.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- McCormack ME, Lopez JA, Crocker TA, Mukhtar MS. Making the right connections: network biology and plant immune system dynamics. Curr. Plant Biol. 2016;5:2–12. doi: 10.1016/j.cpb.2015.10.002. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

