Lipid vesicles chaperone an encapsulated RNA aptamer
- PMID: 29899431
- PMCID: PMC5998061
- DOI: 10.1038/s41467-018-04783-8
Lipid vesicles chaperone an encapsulated RNA aptamer
Abstract
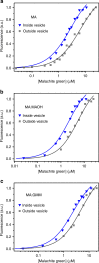
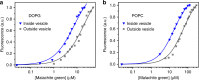
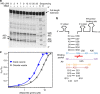
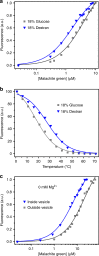
The organization of molecules into cells is believed to have been critical for the emergence of living systems. Early protocells likely consisted of RNA functioning inside vesicles made of simple lipids. However, little is known about how encapsulation would affect the activity and folding of RNA. Here we find that confinement of the malachite green RNA aptamer inside fatty acid vesicles increases binding affinity and locally stabilizes the bound conformation of the RNA. The vesicle effectively 'chaperones' the aptamer, consistent with an excluded volume mechanism due to confinement. Protocellular organization thereby leads to a direct benefit for the RNA. Coupled with previously described mechanisms by which encapsulated RNA aids membrane growth, this effect illustrates how the membrane and RNA might cooperate for mutual benefit. Encapsulation could thus increase RNA fitness and the likelihood that functional sequences would emerge during the origin of life.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Atkins, J., Gesteland, R. & Cech, T. RNA Worlds: From Life’s Origins to Diversity in Gene Regulation (Cold Spring Harbor Laboratory Press, New York, 2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

