Induction and transcriptional regulation of the co-inhibitory gene module in T cells
- PMID: 29899446
- PMCID: PMC6130914
- DOI: 10.1038/s41586-018-0206-z
Induction and transcriptional regulation of the co-inhibitory gene module in T cells
Abstract
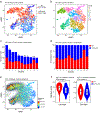
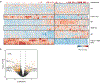
The expression of co-inhibitory receptors, such as CTLA-4 and PD-1, on effector T cells is a key mechanism for ensuring immune homeostasis. Dysregulated expression of co-inhibitory receptors on CD4+ T cells promotes autoimmunity, whereas sustained overexpression on CD8+ T cells promotes T cell dysfunction or exhaustion, leading to impaired ability to clear chronic viral infections and diseases such as cancer1,2. Here, using RNA and protein expression profiling at single-cell resolution in mouse cells, we identify a module of co-inhibitory receptors that includes not only several known co-inhibitory receptors (PD-1, TIM-3, LAG-3 and TIGIT) but also many new surface receptors. We functionally validated two new co-inhibitory receptors, activated protein C receptor (PROCR) and podoplanin (PDPN). The module of co-inhibitory receptors is co-expressed in both CD4+ and CD8+ T cells and is part of a larger co-inhibitory gene program that is shared by non-responsive T cells in several physiological contexts and is driven by the immunoregulatory cytokine IL-27. Computational analysis identified the transcription factors PRDM1 and c-MAF as cooperative regulators of the co-inhibitory module, and this was validated experimentally. This molecular circuit underlies the co-expression of co-inhibitory receptors in T cells and identifies regulators of T cell function with the potential to control autoimmunity and tumour immunity.
Conflict of interest statement
Conflict of Interest
A.C.A. is a member of the SAB for Potenza Therapeutics and Tizona Therapeutics. V.K.K. has an ownership interest and is a member of the SAB for Potenza Therapeutics and Tizona Therapeutics. A.C.A.’s and V.K.K.’s interests were reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict of interest policies. A.R. is an SAB member for Thermo Fisher and Syros Pharmaceuticals and is a consultant for Driver Group.
Figures










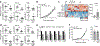

References
-
- Maaten L HG Visualizing Data using t-SNE. Journal of Machine Learning Research, 2579–2605 (2008).
References for Method and Extended Data
-
- Blondel VD, Guillaume JL, Lambiotte R & Lefebvre E Fast unfolding of communities in large networks. J. Stat. Mech, P10008 (2008).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

