Endospores and other lysis-resistant bacteria comprise a widely shared core community within the human microbiota
- PMID: 29899513
- PMCID: PMC6157131
- DOI: 10.1038/s41396-018-0192-z
Endospores and other lysis-resistant bacteria comprise a widely shared core community within the human microbiota
Abstract
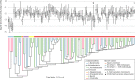
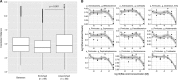
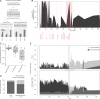
Endospore-formers in the human microbiota are well adapted for host-to-host transmission, and an emerging consensus points to their role in determining health and disease states in the gut. The human gut, more than any other environment, encourages the maintenance of endospore formation, with recent culture-based work suggesting that over 50% of genera in the microbiome carry genes attributed to this trait. However, there has been limited work on the ecological role of endospores and other stress-resistant cellular states in the human gut. In fact, there is no data to indicate whether organisms with the genetic potential to form endospores actually form endospores in situ and how sporulation varies across individuals and over time. Here we applied a culture-independent protocol to enrich for endospores and other stress-resistant cells in human feces to identify variation in these states across people and within an individual over time. We see that cells with resistant states are more likely than those without to be shared among multiple individuals, which suggests that these resistant states are particularly adapted for cross-host dissemination. Furthermore, we use untargeted fecal metabolomics in 24 individuals and within a person over time to show that these organisms respond to shared environmental signals, and in particular, dietary fatty acids, that likely mediate colonization of recently disturbed human guts.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

