UBXN3B positively regulates STING-mediated antiviral immune responses
- PMID: 29899553
- PMCID: PMC5998066
- DOI: 10.1038/s41467-018-04759-8
UBXN3B positively regulates STING-mediated antiviral immune responses
Abstract
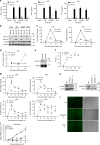
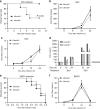
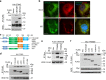
The ubiquitin regulatory X domain-containing proteins (UBXNs) are likely involved in diverse biological processes. Their physiological functions, however, remain largely unknown. Here we present physiological evidence that UBXN3B positively regulates stimulator-of-interferon genes (STING) signaling. We employ a tamoxifen-inducible Cre-LoxP approach to generate systemic Ubxn3b knockout in adult mice as the Ubxn3b-null mutation is embryonically lethal. Ubxn3b-/-, like Sting-/- mice, are highly susceptible to lethal herpes simplex virus 1 (HSV-1) and vesicular stomatitis virus (VSV) infection, which is correlated with deficient immune responses when compared to Ubxn3b+/+ littermates. HSV-1 and STING agonist-induced immune responses are also reduced in several mouse and human Ubxn3b-/- primary cells. Mechanistic studies demonstrate that UBXN3B interacts with both STING and its E3 ligase TRIM56, and facilitates STING ubiquitination, dimerization, trafficking, and consequent recruitment and phosphorylation of TBK1. These results provide physiological evidence that links the UBXN family with antiviral immune responses.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

