CHL1 gene acts as a tumor suppressor in human neuroblastoma
- PMID: 29899830
- PMCID: PMC5995240
- DOI: 10.18632/oncotarget.25403
CHL1 gene acts as a tumor suppressor in human neuroblastoma
Abstract
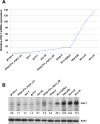
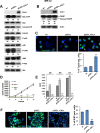
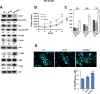
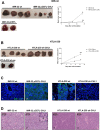
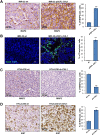
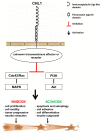
Neuroblastoma is an aggressive, relapse-prone childhood tumor of the sympathetic nervous system that accounts for 15% of pediatric cancer deaths. A distal portion of human chromosome 3p is often deleted in neuroblastoma, this region may contain one or more putative tumor suppressor genes. A 2.54 Mb region at 3p26.3 encompassing the smallest region of deletion pinpointed CHL1 gene, the locus for neuronal cell adhesion molecule close homolog of L1. We found that low CHL1 expression predicted poor outcome in neuroblastoma patients. Here we have used two inducible cell models to analyze the impact of CHL1 on neuroblastoma biology. Over-expression of CHL1 induced neurite-like outgrowth and markers of neuronal differentiation in neuroblastoma cells, halted tumor progression, inhibited anchorage-independent colony formation, and suppressed the growth of human tumor xenografts. Conversely, knock-down of CHL1 induced neurite retraction and activation of Rho GTPases, enhanced cell proliferation and migration, triggered colony formation and anchorage-independent growth, accelerated growth in orthotopic xenografts mouse model. Our findings demonstrate unambiguously that CHL1 acts as a regulator of proliferation and differentiation of neuroblastoma cells through inhibition of the MAPKs and Akt pathways. CHL1 is a novel candidate tumor suppressor in neuroblastoma, and its associated pathways may represent a promising target for future therapeutic interventions.
Keywords: CHL1 gene; apoptosis; autophagy; differentiation; neuroblastoma.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures




References
-
- Spitz R, Hero B, Ernestus K, Berthold F. Deletions in chromosome arms 3p and 11q are new prognostic markers in localized and 4s neuroblastoma. Clin Cancer Res. 2003;9:52–58. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

