Comprehensive mass spectrometry based biomarker discovery and validation platform as applied to diabetic kidney disease
- PMID: 29900119
- PMCID: PMC5988498
- DOI: 10.1016/j.euprot.2016.12.001
Comprehensive mass spectrometry based biomarker discovery and validation platform as applied to diabetic kidney disease
Abstract
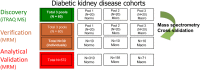
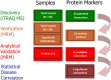
A protein biomarker discovery workflow was applied to plasma samples from patients at different stages of diabetic kidney disease. The proteomics platform produced a panel of significant plasma biomarkers that were statistically scrutinised against the current gold standard tests on an analysis of 572 patients. Five proteins were significantly associated with diabetic kidney disease defined by albuminuria, renal impairment (eGFR) and chronic kidney disease staging (CKD Stage ≥1, ROC curve of 0.77). The results prove the suitability and efficacy of the process used, and introduce a biomarker panel with the potential to improve diagnosis of diabetic kidney disease.
Keywords: Biomarker; Diabetes; Diabetic kidney disease; MRM; iTRAQ.
Figures



References
-
- Liotta L.A., Ferrari M., Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. - PubMed
-
- Ray S., Reddy P.J., Jain R., Gollapalli K., Moiyadi A., Srivastava S. Proteomic technologies for the identification of disease biomarkers in serum: advances and challenges ahead. Proteomics. 2011;11(11):2139–2161. - PubMed
-
- ABSciex technote avaialble at http://www.absciex.jp/Documents/Downloads/Literature/Biomarker-Pipeline-....
-
- Centers for Disease Control and Prevention . U.S. Department of Health and Human Services; Atlanta, GA: 2014. National Chronic Kidney Disease Fact Sheet.http://www.cdc.gov/diabetes/pubs/pdf/kidney_factsheet.pdf
-
- Couser W.G., Remuzzi G., Mendis S., Tonelli M. The contribution of chronic kidney disease to the global burden of major noncommunicable diseases. Kidney Int. 2011;80(December (12)):1258–1270. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

