Mechanisms of lysosomal positioning and movement
- PMID: 29900632
- PMCID: PMC6175085
- DOI: 10.1111/tra.12587
Mechanisms of lysosomal positioning and movement
Abstract
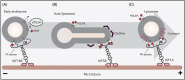
Lysosomes are highly dynamic organelles that can move rapidly throughout the cell. They distribute in a rather immobile pool located around the microtubule-organizing center in a "cloud," and a highly dynamic pool in the cell periphery. Their spatiotemporal characteristics allow them to carry out multiple biological functions, such as cargo degradation, antigen presentation and plasma membrane repair. Therefore, it is not surprising that lysosomal dysfunction underlies various diseases, including cancer, neurodegenerative and autoimmune diseases. In most of these biological events, the involvement of lysosomes is dependent on their ability to move throughout the cytoplasm, to find and fuse to the correct compartments to receive and deliver substrates for further handling. These dynamics are orchestrated by motor proteins moving along cytoskeletal components. The complexity of the mechanisms responsible for controlling lysosomal transport has recently been appreciated and has yielded novel insights into interorganellar communication, as well as lipid-protein interplay. In this review, we discuss the current understanding of the mechanisms of lysosomal transport and the molecular machineries that control this mobility.
Keywords: Arl GTPases; ORP1L; RNF26; Rab GTPases; TOLLIP; cholesterol; dendritic cells; dynein; endoplasmic reticulum; kinesins; late endosome; lysosome; mTOR; membrane contact sites; myosins; phosphatidylinositol phosphate.
© 2018 The Authors. Traffic published by John Wiley & Sons Ltd.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

