Expression of vitamin D receptor in bronchial asthma and its bioinformatics prediction
- PMID: 29901144
- PMCID: PMC6072178
- DOI: 10.3892/mmr.2018.9157
Expression of vitamin D receptor in bronchial asthma and its bioinformatics prediction
Abstract
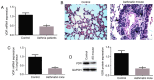
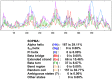
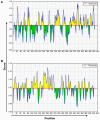
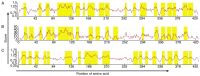
Vitamin D receptors (VDRs) are associated with the occurrence and development of asthma. The aim of the present study was to analyze the secondary structure and B‑cell and T‑cell epitopes of VDR using online prediction software and aid in the future development of a highly efficient epitope‑based vaccine against asthma. Blood samples were collected from peripheral blood of asthmatic children. Reverse transcription quantitative‑polymerase chain reaction (RT‑qPCR) was performed to detect the expression of VDR in the peripheral blood. Mouse models of asthma were established. Hematoxylin and eosin staining was performed to observe the pathological alterations of the lungs of mice. Immunohistochemistry, western blot analysis and RT‑qPCR were performed to detect the expression of VDR in the lungs of asthmatic mice. Online prediction software immune epitope database and analysis resource, SYFPEITHI and linear epitope prediction based on propensity scale and support vector machines were used to predict the B‑cell and T‑cell epitopes and the RasMol and 3DLigandSite were used to analyze the tertiary structure of VDR. RT‑qPCR demonstrated that VDR expression in the peripheral blood of asthmatic children was decreased. Immunohistochemistry, western blotting and RT‑qPCR demonstrated that VDR expression also decreased in the lungs of mouse models of asthma. VDR B‑cell epitopes were identified at 37‑45, 88‑94, 123‑131, 231‑239, 286‑294 and 342‑350 positions of the amino acid sequence and VDR T‑cell epitopes were identified at 125‑130, 231‑239 and 265‑272 positions. A total of six B‑cell epitopes and three T‑cell epitopes for VDR were predicted by bioinformatics, which when validated, may in the future aid in immunological diagnosis and development of a targeted drug therapy for clinical asthma.
Figures




Similar articles
-
Homology modeling and prediction of B‑cell and T‑cell epitopes of the house dust mite allergen Der f 20.Mol Med Rep. 2018 Jan;17(1):1807-1812. doi: 10.3892/mmr.2017.8066. Epub 2017 Nov 15. Mol Med Rep. 2018. PMID: 29257224
-
Bioinformatic prediction of the antigenic epitopes of recombinant ferritin of Echinococcus granulosus.Mol Med Rep. 2016 Jan;13(1):888-94. doi: 10.3892/mmr.2015.4575. Epub 2015 Nov 19. Mol Med Rep. 2016. PMID: 26648019
-
The bioinformatics analyses reveal novel antigen epitopes in major outer membrane protein of Chlamydia trachomatis.Indian J Med Microbiol. 2017 Oct-Dec;35(4):522-528. doi: 10.4103/ijmm.IJMM_17_251. Indian J Med Microbiol. 2017. PMID: 29405144
-
Lessons Learned from Hereditary 1,25-Dihydroxyvitamin D-Resistant Rickets Patients on Vitamin D Functions.J Nutr. 2021 Mar 11;151(3):473-481. doi: 10.1093/jn/nxaa380. J Nutr. 2021. PMID: 33438017 Review.
-
Immunoinformatics and Epitope Prediction.Methods Mol Biol. 2020;2131:155-171. doi: 10.1007/978-1-0716-0389-5_6. Methods Mol Biol. 2020. PMID: 32162252 Review.
References
-
- Vora AC. Bronchial asthma. J Assoc Physicians India. 2014;62(3 Suppl):S5–S6. - PubMed
-
- Kay AB. Mediators of hypersensitivity and inflammatory cells in the pathogenesis of bronchial asthma. Eur J Respir Dis Suppl. 1983;129:1–44. - PubMed
-
- Kuna P. Contemporary views on the pathological mechanism of asthma. Pol Merkur Lekarski. 2003;14:519–521. (In Polish) - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

