Amplification of bacterial genomic DNA from all ascitic fluids with a highly sensitive polymerase chain reaction
- PMID: 29901148
- PMCID: PMC6072169
- DOI: 10.3892/mmr.2018.9159
Amplification of bacterial genomic DNA from all ascitic fluids with a highly sensitive polymerase chain reaction
Abstract
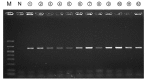
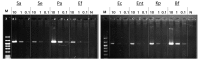
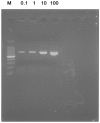
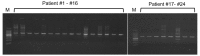
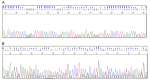
Due to varying positive rates of polymerase chain reaction (PCR) amplification, interpretation of conventional PCR results for non‑infectious ascites remains problematic. The present study developed a highly sensitive PCR protocol and investigated the positive rate of PCR for the 16S ribosomal (r)RNA gene in non‑infectious ascites. Following the design of a new PCR primer pair for the 16S rRNA gene (800F and 1400R), the sequences of PCR products were analyzed and the lower limit for bacterial DNA detection evaluated. The positive rate of PCR for 16S rRNA gene in non‑infectious ascites was also evaluated. PCR with the primer pair amplified the genomic DNA of 16S rRNA genes of major disease‑causing bacterial strains. Additionally, PCR with this primer pair provided highly sensitive detection of bacterial genomic DNA (lower limit, 0.1 pg of template DNA). When DNA samples isolated from ascites were used, the 16S rRNA gene was amplified independently of the presence of bacterial infection. PCR products contained the genomic DNA fragments of multiple bacterial species. Bacterial genomic DNA can be amplified from all ascitic fluids using a highly sensitive PCR protocol. Careful attention is required to interpret the results based on simple amplification of 16S rRNA gene with conventional PCR.
Figures


References
-
- Garcia-Tsao G, Lim JK. Members of Veterans Affairs Hepatitis C Resource Center Program: Management and treatment of patients with cirrhosis and portal hypertension: recommendations from the Department of Veterans Affairs Hepatitis C Resource Center Program and the National Hepatitis C Program. Am J Gastroenterol. 2009;104:1802–1829. doi: 10.1038/ajg.2009.360. - DOI - PubMed
-
- Enomoto H, Inoue S, Matsuhisa A, Aizawa N, Imanishi H, Saito M, Iwata Y, Tanaka H, Ikeda N, Sakai Y, et al. Development of a new in situ hybridization method for the detection of global bacterial DNA to provide early evidence of a bacterial infection in spontaneous bacterial peritonitis. J Hepatol. 2012;56:85–94. doi: 10.1016/j.jhep.2011.06.025. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

