Dynamics of human protein kinase Aurora A linked to drug selectivity
- PMID: 29901437
- PMCID: PMC6054532
- DOI: 10.7554/eLife.36656
Dynamics of human protein kinase Aurora A linked to drug selectivity
Abstract
Protein kinases are major drug targets, but the development of highly-selective inhibitors has been challenging due to the similarity of their active sites. The observation of distinct structural states of the fully-conserved Asp-Phe-Gly (DFG) loop has put the concept of conformational selection for the DFG-state at the center of kinase drug discovery. Recently, it was shown that Gleevec selectivity for the Tyr-kinase Abl was instead rooted in conformational changes after drug binding. Here, we investigate whether protein dynamics after binding is a more general paradigm for drug selectivity by characterizing the binding of several approved drugs to the Ser/Thr-kinase Aurora A. Using a combination of biophysical techniques, we propose a universal drug-binding mechanism, that rationalizes selectivity, affinity and long on-target residence time for kinase inhibitors. These new concepts, where protein dynamics in the drug-bound state plays the crucial role, can be applied to inhibitor design of targets outside the kinome.
Keywords: E. coli; cancer biology; drug binding; enzyme kinetics; molecular biophysics; protein kinase; structural biology.
© 2018, Pitsawong et al.
Conflict of interest statement
WP, VB, RO, RA, AZ, NK, SK, GK, RP, XM, DK No competing interests declared
Figures






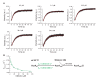





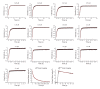


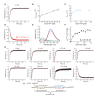

References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

