Patient HLA Germline Variation and Transplant Survivorship
- PMID: 29902106
- PMCID: PMC6097831
- DOI: 10.1200/JCO.2017.77.6534
Patient HLA Germline Variation and Transplant Survivorship
Abstract
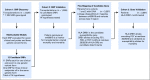
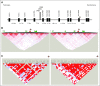
Purpose HLA mismatching increases mortality after unrelated donor hematopoietic cell transplantation. The role of the patient's germline variation on survival is not known. Patients and Methods We previously identified 12 single nucleotide polymorphisms within the HLA region as markers of transplantation determinants and tested these in an independent cohort of 1,555 HLA-mismatched unrelated transplants. Linkage disequilibrium mapping across class II identified candidate susceptibility features. The candidate gene was confirmed in an independent cohort of 3,061 patients. Results Patient rs429916AA/AC was associated with increased transplantation-related mortality compared with rs429916CC (hazard ratio [HR], 1.39; 95% CI, 1.12 to 1.73; P = .003); rs429916A positivity was a proxy for DOA*01:01:05. Mortality increased with one (HR, 1.17; 95% CI, 1.0 to 1.36; P = .05) and two (HR, 2.51; 95% CI, 1.41 to 4.45; P = .002) DOA*01:01:05 alleles. HLA-DOA*01:01:05 was a proxy for HLA-DRB1 alleles encoding FEY ( P < 10E-15) and FDH ( P < 10E-15) amino acid substitutions at residues 26/28/30 that influence HLA-DRβ peptide repertoire. FEY- and FDH-positive alleles were positively associated with rs429916A ( P < 10E-15); FDY-positive alleles were negatively associated. Mortality was increased with FEY (HR, 1.66; 95% CI, 1.29 to 2.13; P = .00008) and FDH (HR, 1.40; 95% CI, 1.02 to 1.93; P = .04), whereas FDY was protective (HR, 0.88; 95% CI, 0.78 to 0.98; P = .02). Of the three candidate motifs, FEY was validated as the susceptibility determinant for mortality (HR, 1.29; 95% CI, 1.00 to 1.67; P = .05). Although FEY was found frequently among African and Hispanic Americans, it increased mortality independently of ancestry. Conclusion Patient germline HLA-DRB1 alleles that encode amino acid substitutions that influence the peptide repertoire of HLA-DRβ predispose to increased death after transplantation. Patient germline variation informs transplantation outcomes across US populations and may provide a means to reduce risks for high-risk patients through pretransplantation screening and evaluation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

