Harnessing Tumor Evolution to Circumvent Resistance
- PMID: 29903534
- PMCID: PMC6368975
- DOI: 10.1016/j.tig.2018.05.007
Harnessing Tumor Evolution to Circumvent Resistance
Abstract
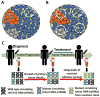
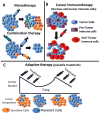
High-throughput sequencing can be used to measure changes in tumor composition across space and time. Specifically, comparisons of pre- and post-treatment samples can reveal the underlying clonal dynamics and resistance mechanisms. Here, we discuss evidence for distinct modes of tumor evolution and their implications for therapeutic strategies. In addition, we consider the utility of spatial tissue sampling schemes, single-cell analysis, and circulating tumor DNA to track tumor evolution and the emergence of resistance, as well as approaches that seek to forestall resistance by targeting tumor evolution. Ultimately, characterization of the (epi)genomic, transcriptomic, and phenotypic changes that occur during tumor progression coupled with computational and mathematical modeling of tumor evolutionary dynamics may inform personalized treatment strategies.
Keywords: clonal dynamics; computational modeling; therapeutic resistance; tissue correlative studies; tumor evolution.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

References
-
- Scott A, et al. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. - PubMed
-
- Ledermann J, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. - PubMed
-
- Druker B, et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2006;359:1757–1765. - PubMed
-
- Holohan C, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

