Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins
- PMID: 29903879
- PMCID: PMC6192670
- DOI: 10.1158/2159-8290.CD-17-1371
Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins
Abstract
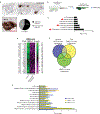
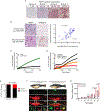
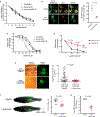
Advanced, metastatic melanomas frequently grow in subcutaneous tissues and portend a poor prognosis. Though subcutaneous tissues are largely composed of adipocytes, the mechanisms by which adipocytes influence melanoma are poorly understood. Using in vitro and in vivo models, we find that adipocytes increase proliferation and invasion of adjacent melanoma cells. Additionally, adipocytes directly transfer lipids to melanoma cells, which alters tumor cell metabolism. Adipocyte-derived lipids are transferred to melanoma cells through the FATP/SLC27A family of lipid transporters expressed on the tumor cell surface. Among the six FATP/SLC27A family members, melanomas significantly overexpress FATP1/SLC27A1. Melanocyte-specific FATP1 expression cooperates with BRAFV600E in transgenic zebrafish to accelerate melanoma development, an effect that is similarly seen in mouse xenograft studies. Pharmacologic blockade of FATPs with the small-molecule inhibitor Lipofermata abrogates lipid transport into melanoma cells and reduces melanoma growth and invasion. These data demonstrate that stromal adipocytes can drive melanoma progression through FATP lipid transporters and represent a new target aimed at interrupting adipocyte-melanoma cross-talk.Significance: We demonstrate that stromal adipocytes are donors of lipids that mediate melanoma progression. Adipocyte-derived lipids are taken up by FATP proteins that are aberrantly expressed in melanoma. Inhibition of FATPs decreases melanoma lipid uptake, invasion, and growth. We provide a mechanism for how stromal adipocytes drive tumor progression and demonstrate a novel microenvironmental therapeutic target. Cancer Discov; 8(8); 1006-25. ©2018 AACR.This article is highlighted in the In This Issue feature, p. 899.
©2018 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest statement:
The authors declare no conflict of interest.
Competing Financial Interests
The authors declare no competing financial interests.
Figures




Comment in
-
Sharing Is Caring.Dev Cell. 2019 May 6;49(3):306-307. doi: 10.1016/j.devcel.2019.04.023. Dev Cell. 2019. PMID: 31063747
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

