Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia
- PMID: 29904051
- PMCID: PMC6002414
- DOI: 10.1038/s41467-018-04737-0
Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia
Abstract
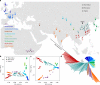
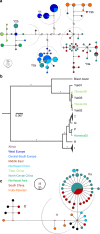
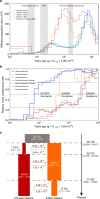
Cattle domestication and the complex histories of East Asian cattle breeds warrant further investigation. Through analysing the genomes of 49 modern breeds and eight East Asian ancient samples, worldwide cattle are consistently classified into five continental groups based on Y-chromosome haplotypes and autosomal variants. We find that East Asian cattle populations are mainly composed of three distinct ancestries, including an earlier East Asian taurine ancestry that reached China at least ~3.9 kya, a later introduced Eurasian taurine ancestry, and a novel Chinese indicine ancestry that diverged from Indian indicine approximately 36.6-49.6 kya. We also report historic introgression events that helped domestic cattle from southern China and the Tibetan Plateau achieve rapid adaptation by acquiring ~2.93% and ~1.22% of their genomes from banteng and yak, respectively. Our findings provide new insights into the evolutionary history of cattle and the importance of introgression in adaptation of cattle to new environmental challenges in East Asia.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- National Bureau of Statistics. China Statistical Yearbook 2016 (China Statistics Press, Beijing, 2016).
-
- Vigne JD, Peters J, Helmer D. The First Steps of Animal Domestication. Oxford: Oxbow Books; 2005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

