LncRNA expression profiling of BMSCs in osteonecrosis of the femoral head associated with increased adipogenic and decreased osteogenic differentiation
- PMID: 29904151
- PMCID: PMC6002551
- DOI: 10.1038/s41598-018-27501-2
LncRNA expression profiling of BMSCs in osteonecrosis of the femoral head associated with increased adipogenic and decreased osteogenic differentiation
Abstract
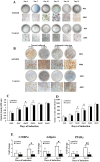
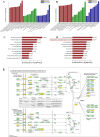
Long noncoding RNAs (lncRNAs) are critical gene expression regulators and are involved in several bone diseases. To explore the potential roles of lncRNAs in osteonecrosis of the femoral head (ONFH), we investigated for the first time the lncRNA expression profile of bone marrow mesenchymal stem cells (BMSCs) from patients with steroid-induced ONFH (SONFH) with microarray and bioinformatics analysis. A total of 1878 lncRNAs and 838 mRNAs were significantly up-regulated while 1842 lncRNAs and 1937 mRNAs were statistically down-regulated in the SONFH group compared with control group. The results validated by qRT-PCR were consistent with the microarray profiling data, especially involved in upregulation and downregulation of critical lncRNAs as well as mRNAs expression related to adipogenic and osteogenic differentiation. Pathway analyses revealed 40 signaling pathways with significant differences, especially the signaling pathways to regulate stem cell pluripotency. The CNC and ceRNA network indicated that lncRNA RP1-193H18.2, MALAT1 and HOTAIR were associated with abnormal osteogenic and adipogenic differentiation of BMSCs in the patients with SONFH. Our results suggest the lncRNA expression profiles were closely associated with the abnormal adipogenic and osteogenic transdifferentiation of BMSCs during the development of SONFH and explore a new insight into the molecular mechanisms of SONFH.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

