The Influence of Higher-Order Epistasis on Biological Fitness Landscape Topography
- PMID: 29904213
- PMCID: PMC5986866
- DOI: 10.1007/s10955-018-1975-3
The Influence of Higher-Order Epistasis on Biological Fitness Landscape Topography
Abstract
The effect of a mutation on the organism often depends on what other mutations are already present in its genome. Geneticists refer to such mutational interactions as epistasis. Pairwise epistatic effects have been recognized for over a century, and their evolutionary implications have received theoretical attention for nearly as long. However, pairwise epistatic interactions themselves can vary with genomic background. This is called higher-order epistasis, and its consequences for evolution are much less well understood. Here, we assess the influence that higher-order epistasis has on the topography of 16 published, biological fitness landscapes. We find that on average, their effects on fitness landscape declines with order, and suggest that notable exceptions to this trend may deserve experimental scrutiny. We conclude by highlighting opportunities for further theoretical and experimental work dissecting the influence that epistasis of all orders has on fitness landscape topography and on the efficiency of evolution by natural selection.
Keywords: Fitness landscapes topography; Higher-order epistasis; NK landscape; Natural selection; Sequence space combinatorics.
Figures

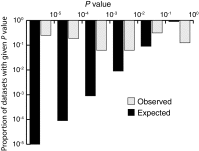
Similar articles
-
Should evolutionary geneticists worry about higher-order epistasis?Curr Opin Genet Dev. 2013 Dec;23(6):700-7. doi: 10.1016/j.gde.2013.10.007. Epub 2013 Nov 27. Curr Opin Genet Dev. 2013. PMID: 24290990 Free PMC article. Review.
-
Analysis of epistatic interactions and fitness landscapes using a new geometric approach.BMC Evol Biol. 2007 Apr 13;7:60. doi: 10.1186/1471-2148-7-60. BMC Evol Biol. 2007. PMID: 17433106 Free PMC article.
-
Effect of Host Species on Topography of the Fitness Landscape for a Plant RNA Virus.J Virol. 2016 Oct 28;90(22):10160-10169. doi: 10.1128/JVI.01243-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581976 Free PMC article.
-
Measuring epistasis in fitness landscapes: The correlation of fitness effects of mutations.J Theor Biol. 2016 May 7;396:132-43. doi: 10.1016/j.jtbi.2016.01.037. Epub 2016 Feb 20. J Theor Biol. 2016. PMID: 26854875
-
Perspective: Sign epistasis and genetic constraint on evolutionary trajectories.Evolution. 2005 Jun;59(6):1165-74. Evolution. 2005. PMID: 16050094 Review.
Cited by
-
Idiosyncratic epistasis leads to global fitness-correlated trends.Science. 2022 May 6;376(6593):630-635. doi: 10.1126/science.abm4774. Epub 2022 May 5. Science. 2022. PMID: 35511982 Free PMC article.
-
Mutations that enhance evolvability may open doors to faster adaptation.Nat Commun. 2023 Oct 9;14(1):6310. doi: 10.1038/s41467-023-41914-2. Nat Commun. 2023. PMID: 37813831 Free PMC article.
-
Evolutionary constraints in fitness landscapes.Heredity (Edinb). 2018 Nov;121(5):466-481. doi: 10.1038/s41437-018-0110-1. Epub 2018 Jul 11. Heredity (Edinb). 2018. PMID: 29993041 Free PMC article.
-
Inferring a complete genotype-phenotype map from a small number of measured phenotypes.PLoS Comput Biol. 2020 Sep 29;16(9):e1008243. doi: 10.1371/journal.pcbi.1008243. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32991585 Free PMC article.
-
Model genotype-phenotype mappings and the algorithmic structure of evolution.J R Soc Interface. 2019 Nov 29;16(160):20190332. doi: 10.1098/rsif.2019.0332. Epub 2019 Nov 6. J R Soc Interface. 2019. PMID: 31690233 Free PMC article.
References
-
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CE, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
