sRNA-dependent control of curli biosynthesis in Escherichia coli: McaS directs endonucleolytic cleavage of csgD mRNA
- PMID: 29905843
- PMCID: PMC6061853
- DOI: 10.1093/nar/gky479
sRNA-dependent control of curli biosynthesis in Escherichia coli: McaS directs endonucleolytic cleavage of csgD mRNA
Abstract
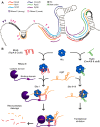
Production of curli, extracellular protein structures important for Escherichia coli biofilm formation, is governed by a highly complex regulatory mechanism that integrates multiple environmental signals through the involvement of numerous proteins and small non-coding RNAs (sRNAs). No less than seven sRNAs (McaS, RprA, GcvB, RydC, RybB, OmrA and OmrB) are known to repress the expression of the curli activator CsgD. Many of the sRNAs repress CsgD production by binding to the csgD mRNA at sites far upstream of the ribosomal binding site. The precise mechanism behind sRNA-mediated regulation of CsgD synthesis is largely unknown. In this study, we identify a conserved A/U-rich region in the csgD mRNA 5' untranslated region, which is cleaved upon binding of the small RNAs, McaS, RprA or GcvB, to sites located more than 30 nucleotides downstream. Mutational analysis shows that the A/U-rich region as well as an adjacent stem-loop structure are required for McaS-stimulated degradation, also serving as a binding platform for the RNA chaperone Hfq. Prevention of McaS-activated cleavage completely relieves repression, suggesting that endoribonucleolytic cleavage of csgD mRNA is the primary regulatory effect exerted by McaS. Moreover, we find that McaS-mediated degradation of the csgD 5' untranslated region requires RNase E.
Figures
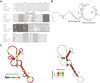
 ) are marked with black arrows and the remaining (
) are marked with black arrows and the remaining (


 ) with grey arrows.
) with grey arrows.  is specific to McaS, while
is specific to McaS, while  is specific to GcvB. McaS, RprA and GcvB all induce cleavage in the conserved A/U-rich region (gray bar).
is specific to GcvB. McaS, RprA and GcvB all induce cleavage in the conserved A/U-rich region (gray bar).

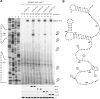
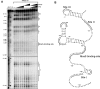
 and
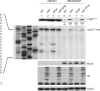
and  from Figure 3 (arrows). (B) Northern blot with RNA purified from SØ928Δ5 carrying either wild-type or mutant pBAD-csgDchiX derivatives (this plasmid contains the csgD 5′UTR and the first 87 nucleotides of the coding region and the small RNA ChiX for stabilisation of the construct) and either pNDM-mcaS or the empty vector pNDM220. Cultures were grown in LB media at 37°C to an OD600 of 0.6 at which point a sample was taken from the culture with wild type csgD and empty pNDM220 vector as a negative control. The cultures were induced with 1 mM IPTG for 10 min, followed by 5 min induction with 0.2% arabinose at which point samples were taken from all cultures. Northern blot was performed as described in methods. (C) Northern and Western blot with RNA and proteins purified from SØ928Δ5 carrying either wild type or mutant pBAD-csgDFLAG and either pNDM-mcaS or the empty vector pNDM220. Cultures were grown and samples were taken as for panel B and Western and northern blots were performed as described in methods. The CsgD-FLAG levels were quantified from three experiments and normalized to the loading control (GroEL). The efficiency of McaS-mediated repression was determined by dividing the signal from samples without McaS by the signal from samples with McaS. Efficiency of McaS on wild type csgD was defined as 1.
from Figure 3 (arrows). (B) Northern blot with RNA purified from SØ928Δ5 carrying either wild-type or mutant pBAD-csgDchiX derivatives (this plasmid contains the csgD 5′UTR and the first 87 nucleotides of the coding region and the small RNA ChiX for stabilisation of the construct) and either pNDM-mcaS or the empty vector pNDM220. Cultures were grown in LB media at 37°C to an OD600 of 0.6 at which point a sample was taken from the culture with wild type csgD and empty pNDM220 vector as a negative control. The cultures were induced with 1 mM IPTG for 10 min, followed by 5 min induction with 0.2% arabinose at which point samples were taken from all cultures. Northern blot was performed as described in methods. (C) Northern and Western blot with RNA and proteins purified from SØ928Δ5 carrying either wild type or mutant pBAD-csgDFLAG and either pNDM-mcaS or the empty vector pNDM220. Cultures were grown and samples were taken as for panel B and Western and northern blots were performed as described in methods. The CsgD-FLAG levels were quantified from three experiments and normalized to the loading control (GroEL). The efficiency of McaS-mediated repression was determined by dividing the signal from samples without McaS by the signal from samples with McaS. Efficiency of McaS on wild type csgD was defined as 1.
References
-
- Flemming H.C., Wingender J.. The biofilm matrix. Nat. Rev. Microbiol. 2010; 8:623–633. - PubMed
-
- Hall-Stoodley L., Costerton J.W., Stoodley P.. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004; 2:95–108. - PubMed
-
- Espeland E.M., Wetzel R.G.. Complexation, stabilization, and UV photolysis of extracellular and surface-bound glucosidase and alkaline phosphatase: implications for biofilm microbiota. Microb. Ecol. 2001; 42:572–585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

