HMMER web server: 2018 update
- PMID: 29905871
- PMCID: PMC6030962
- DOI: 10.1093/nar/gky448
HMMER web server: 2018 update
Abstract
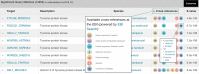
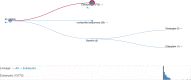
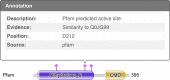
The HMMER webserver [http://www.ebi.ac.uk/Tools/hmmer] is a free-to-use service which provides fast searches against widely used sequence databases and profile hidden Markov model (HMM) libraries using the HMMER software suite (http://hmmer.org). The results of a sequence search may be summarized in a number of ways, allowing users to view and filter the significant hits by domain architecture or taxonomy. For large scale usage, we provide an application programmatic interface (API) which has been expanded in scope, such that all result presentations are available via both HTML and API. Furthermore, we have refactored our JavaScript visualization library to provide standalone components for different result representations. These consume the aforementioned API and can be integrated into third-party websites. The range of databases that can be searched against has been expanded, adding four sequence datasets (12 in total) and one profile HMM library (6 in total). To help users explore the biological context of their results, and to discover new data resources, search results are now supplemented with cross references to other EMBL-EBI databases.
Figures
References
-
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998; 14:755–763. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

