The comparative genomics of Bifidobacterium callitrichos reflects dietary carbohydrate utilization within the common marmoset gut
- PMID: 29906260
- PMCID: PMC6096940
- DOI: 10.1099/mgen.0.000183
The comparative genomics of Bifidobacterium callitrichos reflects dietary carbohydrate utilization within the common marmoset gut
Abstract
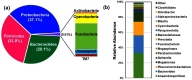
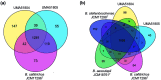
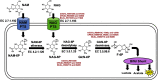
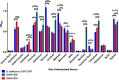
Bifidobacterium is a diverse genus of anaerobic, saccharolytic bacteria that colonize many animals, notably humans and other mammals. The presence of these bacteria in the gastrointestinal tract represents a potential coevolution between the gut microbiome and its mammalian host mediated by diet. To study the relationship between bifidobacterial gut symbionts and host nutrition, we analyzed the genome of two bifidobacteria strains isolated from the feces of a common marmoset (Callithrix jacchus), a primate species studied for its ability to subsist on host-indigestible carbohydrates. Whole genome sequencing identified these isolates as unique strains of Bifidobacterium callitrichos. All three strains, including these isolates and the previously described type strain, contain genes that may enable utilization of marmoset dietary substrates. These include genes predicted to contribute to galactose, arabinose, and trehalose metabolic pathways. In addition, significant genomic differences between strains suggest that bifidobacteria possess distinct roles in carbohydrate metabolism within the same host. Thus, bifidobacteria utilize dietary components specific to their host, both humans and non-human primates alike. Comparative genomics suggests conservation of possible coevolutionary relationships within the primate clade.
Keywords: bifidobacteria; commensalism; comparative genomics; gut microbiota; non-human primate.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Captive Common Marmosets (Callithrix jacchus) Are Colonized throughout Their Lives by a Community of Bifidobacterium Species with Species-Specific Genomic Content That Can Support Adaptation to Distinct Metabolic Niches.mBio. 2021 Aug 31;12(4):e0115321. doi: 10.1128/mBio.01153-21. Epub 2021 Aug 3. mBio. 2021. PMID: 34340536 Free PMC article.
-
Comparative genomics of Bifidobacterium species isolated from marmosets and humans.Am J Primatol. 2019 Oct;81(10-11):e983. doi: 10.1002/ajp.22983. Epub 2019 May 6. Am J Primatol. 2019. PMID: 31062394 Free PMC article.
-
Bifidobacterium reuteri sp. nov., Bifidobacterium callitrichos sp. nov., Bifidobacterium saguini sp. nov., Bifidobacterium stellenboschense sp. nov. and Bifidobacterium biavatii sp. nov. isolated from faeces of common marmoset (Callithrix jacchus) and red-handed tamarin (Saguinus midas).Syst Appl Microbiol. 2012 Mar;35(2):92-7. doi: 10.1016/j.syapm.2011.11.006. Epub 2012 Jan 4. Syst Appl Microbiol. 2012. PMID: 22225994
-
Glycan Utilization and Cross-Feeding Activities by Bifidobacteria.Trends Microbiol. 2018 Apr;26(4):339-350. doi: 10.1016/j.tim.2017.10.001. Epub 2017 Oct 28. Trends Microbiol. 2018. PMID: 29089173 Review.
-
Genomics of the Genus Bifidobacterium Reveals Species-Specific Adaptation to the Glycan-Rich Gut Environment.Appl Environ Microbiol. 2015 Nov 20;82(4):980-991. doi: 10.1128/AEM.03500-15. Print 2016 Feb 15. Appl Environ Microbiol. 2015. PMID: 26590291 Free PMC article. Review.
Cited by
-
Genomics-based analysis of four porcine-derived lactic acid bacteria strains and their evaluation as potential probiotics.Mol Genet Genomics. 2024 Mar 5;299(1):24. doi: 10.1007/s00438-024-02101-0. Mol Genet Genomics. 2024. PMID: 38438804
-
Captive Common Marmosets (Callithrix jacchus) Are Colonized throughout Their Lives by a Community of Bifidobacterium Species with Species-Specific Genomic Content That Can Support Adaptation to Distinct Metabolic Niches.mBio. 2021 Aug 31;12(4):e0115321. doi: 10.1128/mBio.01153-21. Epub 2021 Aug 3. mBio. 2021. PMID: 34340536 Free PMC article.
-
Analysis of gut microbiome profiles in common marmosets (Callithrix jacchus) in health and intestinal disease.Sci Rep. 2022 Mar 15;12(1):4430. doi: 10.1038/s41598-022-08255-4. Sci Rep. 2022. PMID: 35292670 Free PMC article.
-
Recent Advances in the Etiology, Diagnosis, and Treatment of Marmoset Wasting Syndrome.Vet Sci. 2025 Feb 27;12(3):203. doi: 10.3390/vetsci12030203. Vet Sci. 2025. PMID: 40266930 Free PMC article. Review.
-
Metabolomics reveals impact of seven functional foods on metabolic pathways in a gut microbiota model.J Adv Res. 2020 Jan 3;23:47-59. doi: 10.1016/j.jare.2020.01.001. eCollection 2020 May. J Adv Res. 2020. PMID: 32071791 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

