Prospectively Isolated Tetraspanin+ Neoblasts Are Adult Pluripotent Stem Cells Underlying Planaria Regeneration
- PMID: 29906446
- PMCID: PMC9359418
- DOI: 10.1016/j.cell.2018.05.006
Prospectively Isolated Tetraspanin+ Neoblasts Are Adult Pluripotent Stem Cells Underlying Planaria Regeneration
Abstract
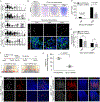
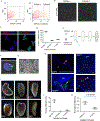
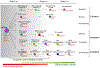
Proliferating cells known as neoblasts include pluripotent stem cells (PSCs) that sustain tissue homeostasis and regeneration of lost body parts in planarians. However, the lack of markers to prospectively identify and isolate these adult PSCs has significantly hampered their characterization. We used single-cell RNA sequencing (scRNA-seq) and single-cell transplantation to address this long-standing issue. Large-scale scRNA-seq of sorted neoblasts unveiled a novel subtype of neoblast (Nb2) characterized by high levels of PIWI-1 mRNA and protein and marked by a conserved cell-surface protein-coding gene, tetraspanin 1 (tspan-1). tspan-1-positive cells survived sub-lethal irradiation, underwent clonal expansion to repopulate whole animals, and when purified with an anti-TSPAN-1 antibody, rescued the viability of lethally irradiated animals after single-cell transplantation. The first prospective isolation of an adult PSC bridges a conceptual dichotomy between functionally and molecularly defined neoblasts, shedding light on mechanisms governing in vivo pluripotency and a source of regeneration in animals. VIDEO ABSTRACT.
Keywords: planaria; pluripotent; prospective isolation; regeneration; single cell transplantation; stem cells; tetraspanin.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




Comment in
-
Prospecting for Planarian Pluripotency.Cell. 2018 Jun 14;173(7):1566-1567. doi: 10.1016/j.cell.2018.05.062. Cell. 2018. PMID: 29906442
-
Prospecting for pluripotency.Nat Rev Genet. 2018 Aug;19(8):471. doi: 10.1038/s41576-018-0030-1. Nat Rev Genet. 2018. PMID: 29934629 No abstract available.
-
Finding the potency in planarians.Commun Biol. 2022 Sep 15;5(1):970. doi: 10.1038/s42003-022-03905-9. Commun Biol. 2022. PMID: 36109651 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

