Predicting human genes susceptible to genomic instability associated with Alu/ Alu-mediated rearrangements
- PMID: 29907612
- PMCID: PMC6071635
- DOI: 10.1101/gr.229401.117
Predicting human genes susceptible to genomic instability associated with Alu/ Alu-mediated rearrangements
Abstract
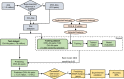
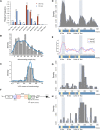
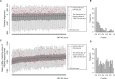
Alu elements, the short interspersed element numbering more than 1 million copies per human genome, can mediate the formation of copy number variants (CNVs) between substrate pairs. These Alu/Alu-mediated rearrangements (AAMRs) can result in pathogenic variants that cause diseases. To investigate the impact of AAMR on gene variation and human health, we first characterized Alus that are involved in mediating CNVs (CNV-Alus) and observed that these Alus tend to be evolutionarily younger. We then computationally generated, with the assistance of a supercomputer, a test data set consisting of 78 million Alu pairs and predicted ∼18% of them are potentially susceptible to AAMR. We further determined the relative risk of AAMR in 12,074 OMIM genes using the count of predicted CNV-Alu pairs and experimentally validated the predictions with 89 samples selected by correlating predicted hotspots with a database of CNVs identified by clinical chromosomal microarrays (CMAs) on the genomes of approximately 54,000 subjects. We fine-mapped 47 duplications, 40 deletions, and two complex rearrangements and examined a total of 52 breakpoint junctions of simple CNVs. Overall, 94% of the candidate breakpoints were at least partially Alu mediated. We successfully predicted all (100%) of Alu pairs that mediated deletions (n = 21) and achieved an 87% positive predictive value overall when including AAMR-generated deletions and duplications. We provided a tool, AluAluCNVpredictor, for assessing AAMR hotspots and their role in human disease. These results demonstrate the utility of our predictive model and provide insights into the genomic features and molecular mechanisms underlying AAMR.
© 2018 Song et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3.Hum Mol Genet. 2015 Jul 15;24(14):4061-77. doi: 10.1093/hmg/ddv146. Epub 2015 Apr 23. Hum Mol Genet. 2015. PMID: 25908615 Free PMC article.
-
The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles.Am J Hum Genet. 2014 Aug 7;95(2):143-61. doi: 10.1016/j.ajhg.2014.06.014. Epub 2014 Jul 24. Am J Hum Genet. 2014. PMID: 25065914 Free PMC article.
-
Human endogenous retroviral elements promote genome instability via non-allelic homologous recombination.BMC Biol. 2014 Sep 23;12:74. doi: 10.1186/s12915-014-0074-4. BMC Biol. 2014. PMID: 25246103 Free PMC article.
-
Alu elements: an intrinsic source of human genome instability.Curr Opin Virol. 2013 Dec;3(6):639-45. doi: 10.1016/j.coviro.2013.09.002. Epub 2013 Sep 27. Curr Opin Virol. 2013. PMID: 24080407 Free PMC article. Review.
-
A Variety of Alu-Mediated Copy Number Variations Can Underlie IL-12Rβ1 Deficiency.J Clin Immunol. 2018 Jul;38(5):617-627. doi: 10.1007/s10875-018-0527-6. Epub 2018 Jul 11. J Clin Immunol. 2018. PMID: 29995221 Free PMC article. Review.
Cited by
-
Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders.Cell. 2022 May 26;185(11):1986-2005.e26. doi: 10.1016/j.cell.2022.04.017. Epub 2022 May 6. Cell. 2022. PMID: 35525246 Free PMC article.
-
Systematic assessment of the contribution of structural variants to inherited retinal diseases.Hum Mol Genet. 2023 Jun 5;32(12):2005-2015. doi: 10.1093/hmg/ddad032. Hum Mol Genet. 2023. PMID: 36811936 Free PMC article.
-
Palindromes in DNA-A Risk for Genome Stability and Implications in Cancer.Int J Mol Sci. 2021 Mar 11;22(6):2840. doi: 10.3390/ijms22062840. Int J Mol Sci. 2021. PMID: 33799581 Free PMC article. Review.
-
CRISPR/Cas9-induced gene conversion between ATAD3 paralogs.HGG Adv. 2022 Jan 25;3(2):100092. doi: 10.1016/j.xhgg.2022.100092. eCollection 2022 Apr 14. HGG Adv. 2022. PMID: 35199044 Free PMC article.
-
Genetic genealogy uncovers a founder deletion mutation in the cerebral cavernous malformations 2 gene.Hum Genet. 2022 Nov;141(11):1761-1769. doi: 10.1007/s00439-022-02458-5. Epub 2022 Apr 30. Hum Genet. 2022. PMID: 35488064 Free PMC article.
References
-
- Batzer MA, Deininger PL. 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
